7THV
 
 | | Structure of the yeast clamp loader (Replication Factor C RFC) bound to the sliding clamp (Proliferating Cell Nuclear Antigen PCNA) in an autoinhibited conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Gaubitz, C, Liu, X, Pajak, J, Stone, N, Hayes, J, Demo, G, Kelch, B.A. | | Deposit date: | 2022-01-12 | | Release date: | 2022-02-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures reveal high-resolution mechanism of a DNA polymerase sliding clamp loader.
Elife, 11, 2022
|
|
7TKU
 
 | | Structure of the yeast clamp loader (Replication Factor C RFC) bound to the open sliding clamp (Proliferating Cell Nuclear Antigen PCNA) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Gaubitz, C, Liu, X, Pajak, J, Stone, N, Hayes, J, Demo, G, Kelch, B.A. | | Deposit date: | 2022-01-17 | | Release date: | 2022-02-16 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures reveal high-resolution mechanism of a DNA polymerase sliding clamp loader.
Elife, 11, 2022
|
|
7TIB
 
 | | Structure of the yeast clamp loader (Replication Factor C RFC) bound to the open sliding clamp (Proliferating Cell Nuclear Antigen PCNA) and primer-template DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(*AP*GP*AP*CP*AP*CP*TP*AP*CP*GP*AP*GP*TP*AP*CP*AP*TP*A)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*AP*TP*GP*TP*AP*CP*TP*CP*GP*TP*AP*GP*TP*GP*TP*CP*T)-3'), ... | | Authors: | Gaubitz, C, Liu, X, Pajak, J, Stone, N, Hayes, J, Demo, G, Kelch, B.A. | | Deposit date: | 2022-01-13 | | Release date: | 2022-02-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures reveal high-resolution mechanism of a DNA polymerase sliding clamp loader.
Elife, 11, 2022
|
|
7TIC
 
 | | Structure of the yeast clamp loader (Replication Factor C RFC) bound to the sliding clamp (Proliferating Cell Nuclear Antigen PCNA) in an autoinhibited conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Gaubitz, C, Liu, X, Pajak, J, Stone, N, Hayes, J, Demo, G, Kelch, B.A. | | Deposit date: | 2022-01-13 | | Release date: | 2022-02-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structures reveal high-resolution mechanism of a DNA polymerase sliding clamp loader.
Elife, 11, 2022
|
|
7TI8
 
 | | Structure of the yeast clamp loader (Replication Factor C RFC) bound to the open sliding clamp (Proliferating Cell Nuclear Antigen PCNA) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Gaubitz, C, Liu, X, Pajak, J, Stone, N, Hayes, J, Demo, G, Kelch, B.A. | | Deposit date: | 2022-01-13 | | Release date: | 2022-02-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures reveal high-resolution mechanism of a DNA polymerase sliding clamp loader.
Elife, 11, 2022
|
|
7TID
 
 | | Structure of the yeast clamp loader (Replication Factor C RFC) bound to the sliding clamp (Proliferating Cell Nuclear Antigen PCNA) and primer-template DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(*AP*GP*AP*CP*AP*CP*TP*AP*CP*GP*AP*GP*TP*AP*CP*AP*TP*A)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*AP*TP*GP*TP*AP*CP*TP*CP*GP*TP*AP*GP*TP*GP*TP*CP*T)-3'), ... | | Authors: | Gaubitz, C, Liu, X, Pajak, J, Stone, N, Hayes, J, Demo, G, Kelch, B.A. | | Deposit date: | 2022-01-13 | | Release date: | 2022-02-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures reveal high-resolution mechanism of a DNA polymerase sliding clamp loader.
Elife, 11, 2022
|
|
4V2T
 
 | | Membrane embedded pleurotolysin pore with 13 fold symmetry | | Descriptor: | PLEUROTOLYSIN A, PLEUROTOLYSIN B | | Authors: | Lukoyanova, N, Kondos, S.C, Farabella, I, Law, R.H.P, Reboul, C.F, Caradoc-Davies, T.T, Spicer, B.A, Kleifeld, O, Perugini, M, Ekkel, S, Hatfaludi, T, Oliver, K, Hotze, E.M, Tweten, R.K, Whisstock, J.C, Topf, M, Dunstone, M.A, Saibil, H.R. | | Deposit date: | 2014-10-15 | | Release date: | 2015-02-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Conformational Changes During Pore Formation by the Perforin-Related Protein Pleurotolysin.
Plos Biol., 13, 2015
|
|
4WUW
 
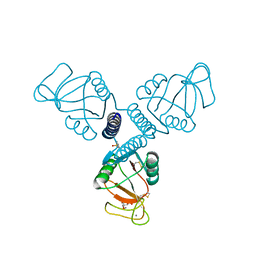 | | Crystal Structure of Surfactant Protein-A DED Mutant (E171D/P175E/K203D) Complexed with Inositol | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, CALCIUM ION, Pulmonary surfactant-associated protein A | | Authors: | Rynkiewicz, M.J, Wu, H, Cafarella, T.R, Nikolaidis, N.M, Head, J.F, Seaton, B.A, McCormack, F.X. | | Deposit date: | 2014-11-03 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.398 Å) | | Cite: | Differential ligand binding specificities of the pulmonary collectins are determined by the conformational freedom of a surface loop
To be published
|
|
4WR9
 
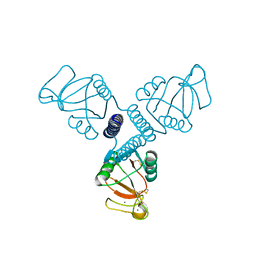 | | Crystal Structure of Surfactant Protein-A DED Mutant (E171D/P175E/K203D) | | Descriptor: | CALCIUM ION, Pulmonary surfactant-associated protein A | | Authors: | Rynkiewicz, M.J, Wu, H, Cafarella, T.R, Nikolaidis, N.M, Head, J.F, Seaton, B.A, McCormack, F.X. | | Deposit date: | 2014-10-23 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Differential ligand binding specificities of the pulmonary collectins are determined by the conformational freedom of a surface loop
To be published
|
|
4WRC
 
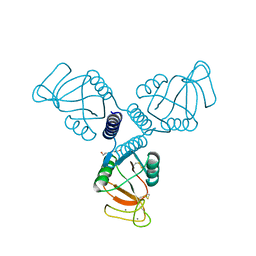 | | Crystal Structure of Surfactant Protein-A DEDN Mutant (E171D/P175E/R197N/K203D) | | Descriptor: | CALCIUM ION, CHLORIDE ION, HEXANE-1,6-DIOL, ... | | Authors: | Rynkiewicz, M.J, Wu, H, Cafarella, T.R, Nikolaidis, N.M, Head, J.F, Seaton, B.A, McCormack, F.X. | | Deposit date: | 2014-10-23 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Differential ligand binding specificities of the pulmonary collectins are determined by the conformational freedom of a surface loop
to be published
|
|
4WRF
 
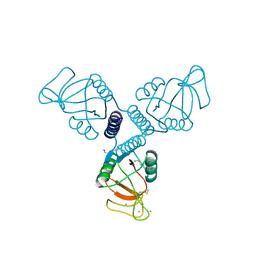 | | Crystal Structure of Surfactant Protein-A DEDN Mutant (E171D/P175E/R197N/K203D) Complexed with Mannose | | Descriptor: | CALCIUM ION, CHLORIDE ION, HEXANE-1,6-DIOL, ... | | Authors: | Rynkiewicz, M.J, Wu, H, Cafarella, T.R, Nikolaidis, N.M, Head, J.F, Seaton, B.A, McCormack, F.X. | | Deposit date: | 2014-10-23 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Differential ligand binding specificities of the pulmonary collectins are determined by the conformational freedom of a surface loop
To be published
|
|
4WRE
 
 | | Crystal Structure of Surfactant Protein-A DEDN Mutant (E171D/P175E/R197N/K203D) Complexed with Inositol | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Rynkiewicz, M.J, Wu, H, Cafarella, T.R, Nikolaidis, N.M, Head, J.F, Seaton, B.A, McCormack, F.X. | | Deposit date: | 2014-10-23 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Differential ligand binding specificities of the pulmonary collectins are determined by the conformational freedom of a surface loop
To be published
|
|
4WUY
 
 | | Crystal Structure of Protein Lysine Methyltransferase SMYD2 in complex with LLY-507, a Cell-Active, Potent and Selective Inhibitor | | Descriptor: | 5-cyano-2'-{4-[2-(3-methyl-1H-indol-1-yl)ethyl]piperazin-1-yl}-N-[3-(pyrrolidin-1-yl)propyl]biphenyl-3-carboxamide, GLYCEROL, N-lysine methyltransferase SMYD2, ... | | Authors: | Nguyen, H, Allali-Hassani, A, Antonysamy, S, Chang, S, Chen, L.H, Curtis, C, Emtage, S, Fan, L, Gheyi, T, Li, F, Liu, S, Martin, J.R, Mendel, D, Olsen, J.B, Pelletier, L, Shatseva, T, Wu, S, Zhang, F.F, Arrowsmith, C.H, Brown, P.J, Campbell, R.M, Garcia, B.A, Barsyte-Lovejoy, D, Mader, M, Vedadi, M. | | Deposit date: | 2014-11-04 | | Release date: | 2015-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | LLY-507, a Cell-active, Potent, and Selective Inhibitor of Protein-lysine Methyltransferase SMYD2.
J.Biol.Chem., 290, 2015
|
|
4WUX
 
 | | Crystal Structure of Surfactant Protein-A DED Mutant (E171D/P175E/K203D) Complexed with Mannose | | Descriptor: | CALCIUM ION, Pulmonary surfactant-associated protein A, alpha-D-mannopyranose | | Authors: | Rynkiewicz, M.J, Wu, H, Cafarella, T.R, Nikolaidis, N.M, Head, J.F, Seaton, B.A, McCormack, F.X. | | Deposit date: | 2014-11-03 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Differential ligand binding specificities of the pulmonary collectins are determined by the conformational freedom of a surface loop
To be published
|
|
6N0Q
 
 | | BRAF in complex with N-(4-methyl-3-(1-methyl-2-oxo-2,3-dihydro-1H-benzo[d]imidazol-5-yl)phenyl)-3-(trifluoromethyl)benzamide. | | Descriptor: | N-[4-methyl-3-(1-methyl-2-oxo-2,3-dihydro-1H-benzimidazol-5-yl)phenyl]-3-(trifluoromethyl)benzamide, Serine/threonine-protein kinase B-raf | | Authors: | Mamo, M, Appleton, B.A. | | Deposit date: | 2018-11-07 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Design and Discovery ofN-(3-(2-(2-Hydroxyethoxy)-6-morpholinopyridin-4-yl)-4-methylphenyl)-2-(trifluoromethyl)isonicotinamide, a Selective, Efficacious, and Well-Tolerated RAF Inhibitor Targeting RAS Mutant Cancers: The Path to the Clinic.
J.Med.Chem., 63, 2020
|
|
6NEU
 
 | | FAD-dependent monooxygenase TropB from T. stipitatus R206Q variant | | Descriptor: | CHLORIDE ION, FAD-dependent monooxygenase tropB, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Rodriguez Benitez, A, Tweedy, S.E, Baker Dockrey, S.A, Lukowski, A.L, Wymore, T, Khare, D, Brooks, C.L, Palfey, B.A, Smith, J.L, Narayan, A.R.H. | | Deposit date: | 2018-12-18 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for selectivity in flavin-dependent monooxygenase-catalyzed oxidative dearomatization.
Acs Catalysis, 9, 2019
|
|
6NEV
 
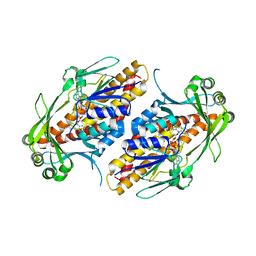 | | FAD-dependent monooxygenase TropB from T. stipitatus Y239F Variant | | Descriptor: | CHLORIDE ION, FAD-dependent monooxygenase tropB, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Rodriguez Benitez, A, Tweedy, S.E, Baker Dockrey, S.A, Lukowski, A.L, Wymore, T, Khare, D, Palfey, B.A, Smith, J.L, Narayan, A.R.H. | | Deposit date: | 2018-12-18 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Structural basis for selectivity in flavin-dependent monooxygenase-catalyzed oxidative dearomatization.
Acs Catalysis, 9, 2019
|
|
6O3H
 
 | | Icosahedral reconstruction of the thermophilic bacteriophage P74-26 capsid | | Descriptor: | Major head protein, P74-26 Head Decoration Protein | | Authors: | Stone, N.P, Demo, G, Agnello, E, Kelch, B.A. | | Deposit date: | 2019-02-26 | | Release date: | 2019-07-24 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Principles for enhancing virus capsid capacity and stability from a thermophilic virus capsid structure.
Nat Commun, 10, 2019
|
|
6NES
 
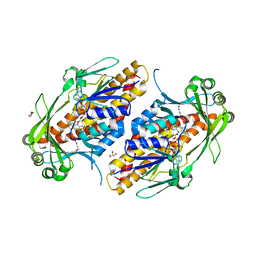 | | FAD-dependent monooxygenase TropB from T. stipitatus | | Descriptor: | CHLORIDE ION, FAD-dependent monooxygenase tropB, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Rodriguez Benitez, A, Tweedy, S.E, Baker Dockrey, S.A, Lukowski, A.L, Wymore, T, Khare, D, Brooks, C.L, Palfey, B.A, Smith, J.L, Narayan, A.R.H. | | Deposit date: | 2018-12-18 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for selectivity in flavin-dependent monooxygenase-catalyzed oxidative dearomatization.
Acs Catalysis, 9, 2019
|
|
6NET
 
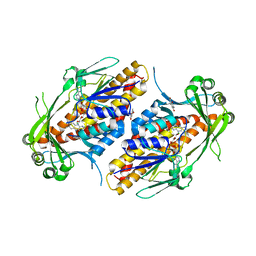 | | FAD-dependent monooxygenase TropB from T. stipitatus substrate complex | | Descriptor: | 2,4-dihydroxy-3,6-dimethylbenzaldehyde, CHLORIDE ION, FAD-dependent monooxygenase tropB, ... | | Authors: | Rodriguez Benitez, A, Tweedy, S.E, Baker Dockrey, S.A, Lukowski, A.L, Wymore, T, Khare, D, Brooks, C.L, Palfey, B.A, Smith, J.L, Narayan, A.R.H. | | Deposit date: | 2018-12-18 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for selectivity in flavin-dependent monooxygenase-catalyzed oxidative dearomatization.
Acs Catalysis, 9, 2019
|
|
6OP6
 
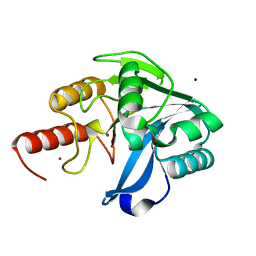 | | Structure of VIM-20 in the reduced state | | Descriptor: | Metallo-beta-lactamase VIM-20, SODIUM ION, ZINC ION | | Authors: | Page, R.C, Shurina, B.A, Montgomery, J.S, Orischak, M.G, Nix, J.C. | | Deposit date: | 2019-04-24 | | Release date: | 2019-10-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | A Single Salt Bridge in VIM-20 Increases Protein Stability and Antibiotic Resistance under Low-Zinc Conditions.
Mbio, 10, 2019
|
|
6N0P
 
 | |
6OJJ
 
 | |
6XKU
 
 | | R. capsulatus cyt bc1 with one FeS protein in b position and one in c position (CIII2 b-c) | | Descriptor: | Cytochrome b, Cytochrome c1, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Steimle, S, Van Eeuwen, T, Ozturk, Y, Kim, H.J, Braitbard, M, Selamoglu, N, Garcia, B.A, Schneidman-Duhovny, D, Murakami, K, Daldal, F. | | Deposit date: | 2020-06-27 | | Release date: | 2020-12-30 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structures of engineered active bc 1 -cbb 3 type CIII 2 CIV super-complexes and electronic communication between the complexes.
Nat Commun, 12, 2021
|
|
6XKZ
 
 | | R. capsulatus CIII2CIV tripartite super-complex, conformation B (SC-1B) | | Descriptor: | COPPER (II) ION, Cbb3-type cytochrome c oxidase subunit CcoP,Cytochrome c-type cyt cy, Cytochrome b, ... | | Authors: | Steimle, S, Van Eeuwen, T, Ozturk, Y, Kim, H.J, Braitbard, M, Selamoglu, N, Garcia, B.A, Schneidman-Duhovny, D, Murakami, K, Daldal, F. | | Deposit date: | 2020-06-27 | | Release date: | 2020-12-30 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Cryo-EM structures of engineered active bc 1 -cbb 3 type CIII 2 CIV super-complexes and electronic communication between the complexes.
Nat Commun, 12, 2021
|
|
