8IFB
 
 | | Dibekacin-bound E.coli 70S ribosome in the PURE system | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Tomono, J, Asano, K, Chiashi, T, Tanaka, Y, Yokoyama, T. | | Deposit date: | 2023-02-17 | | Release date: | 2024-02-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.43 Å) | | Cite: | Direct visualization of ribosomes in the cell-free system revealed the functional evolution of aminoglycoside.
J.Biochem., 175, 2024
|
|
7W3W
 
 | | X-ray structure of apo-VmFbpA, a ferric ion-binding protein from Vibrio metschnikovii | | Descriptor: | Iron-utilization periplasmic protein | | Authors: | Lu, P, Sui, M, Zhang, M, Nagata, K. | | Deposit date: | 2021-11-26 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.858 Å) | | Cite: | Rosmarinic Acid and Sodium Citrate Have a Synergistic Bacteriostatic Effect against Vibrio Species by Inhibiting Iron Uptake.
Int J Mol Sci, 22, 2021
|
|
6L49
 
 | | H3-CA-H3 tri-nucleosome with the 22 base-pair linker DNA | | Descriptor: | DNA (485-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Takizawa, Y, Ho, C.-H, Tachiwana, H, Matsunami, H, Ohi, M, Wolf, M, Kurumizaka, H. | | Deposit date: | 2019-10-16 | | Release date: | 2019-12-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (18.9 Å) | | Cite: | Cryo-EM Structures of Centromeric Tri-nucleosomes Containing a Central CENP-A Nucleosome.
Structure, 28, 2020
|
|
6L4A
 
 | | H3-H3-H3 tri-nucleosome with the 22 base-pair linker DNA | | Descriptor: | DNA (485-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Takizawa, Y, Ho, C.-H, Tachiwana, H, Matsunami, H, Ohi, M, Wolf, M, Kurumizaka, H. | | Deposit date: | 2019-10-16 | | Release date: | 2019-12-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (12.3 Å) | | Cite: | Cryo-EM Structures of Centromeric Tri-nucleosomes Containing a Central CENP-A Nucleosome.
Structure, 28, 2020
|
|
7CJI
 
 | | Photosystem II structure in the S1 state | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Li, H, Shen, J.-R, Suga, M. | | Deposit date: | 2020-07-11 | | Release date: | 2021-04-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Capturing structural changes of the S 1 to S 2 transition of photosystem II using time-resolved serial femtosecond crystallography.
Iucrj, 8, 2021
|
|
7CJJ
 
 | | Photosystem II structure in the S2 state | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Li, H, Shen, J.-R, Suga, M. | | Deposit date: | 2020-07-11 | | Release date: | 2021-04-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Capturing structural changes of the S 1 to S 2 transition of photosystem II using time-resolved serial femtosecond crystallography.
Iucrj, 8, 2021
|
|
7COU
 
 | | Structure of cyanobacterial photosystem II in the dark S1 state | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Li, H, Shen, J.-R, Suga, M. | | Deposit date: | 2020-08-05 | | Release date: | 2021-04-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Capturing structural changes of the S 1 to S 2 transition of photosystem II using time-resolved serial femtosecond crystallography.
Iucrj, 8, 2021
|
|
3W80
 
 | |
3W7Y
 
 | |
1GQV
 
 | | Atomic Resolution (0.98A) Structure of Eosinophil-Derived Neurotoxin | | Descriptor: | ACETATE ION, EOSINOPHIL-DERIVED NEUROTOXIN | | Authors: | Swaminathan, G.J, Holloway, D.E, Veluraja, K, Acharya, K.R. | | Deposit date: | 2001-12-05 | | Release date: | 2002-03-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Atomic Resolution (0.98 A) Structure of Eosinophil-Derived Neurotoxin
Biochemistry, 41, 2002
|
|
7YNH
 
 | |
5ETY
 
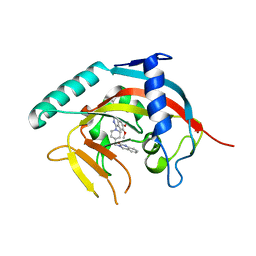 | | Crystal Structure of human Tankyrase-1 bound to K-756 | | Descriptor: | 3-[[1-(6,7-dimethoxyquinazolin-4-yl)piperidin-4-yl]methyl]-1,4-dihydroquinazolin-2-one, Tankyrase-1, ZINC ION | | Authors: | Takahashi, Y, Miyagi, H, Suzuki, M, Saito, J. | | Deposit date: | 2015-11-18 | | Release date: | 2016-06-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Discovery and Characterization of K-756, a Novel Wnt/ beta-Catenin Pathway Inhibitor Targeting Tankyrase
Mol.Cancer Ther., 15, 2016
|
|
8JB7
 
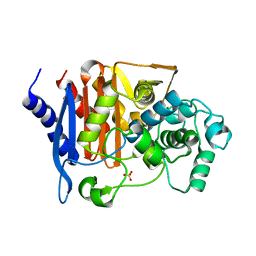 | | Crystal structure of CMY-185 | | Descriptor: | Beta-lactamase, SULFATE ION | | Authors: | Kawai, A, Doi, Y. | | Deposit date: | 2023-05-08 | | Release date: | 2023-12-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural insights into the molecular mechanism of high-level ceftazidime-avibactam resistance conferred by CMY-185.
Mbio, 15, 2024
|
|
8JB8
 
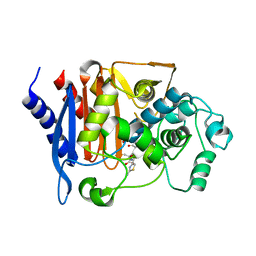 | | Crystal structure of CMY-185 complex with ceftazidime | | Descriptor: | ACYLATED CEFTAZIDIME, Beta-lactamase | | Authors: | Kawai, A, Doi, Y. | | Deposit date: | 2023-05-08 | | Release date: | 2023-12-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into the molecular mechanism of high-level ceftazidime-avibactam resistance conferred by CMY-185.
Mbio, 15, 2024
|
|
8J4H
 
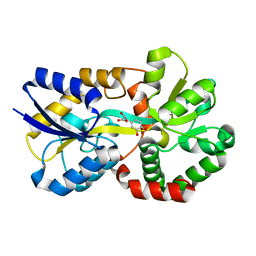 | | X-ray structure of a ferric ion-binding protein A (FbpA) from Vibrio metschnikovii in complex with Danshensu (DSS) | | Descriptor: | (2~{R})-3-[3,4-bis(oxidanyl)phenyl]-2-oxidanyl-propanoic acid, Ferric iron ABC transporter iron-binding protein | | Authors: | Lu, P, Jiang, J, Nagata, K. | | Deposit date: | 2023-04-20 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Molecular mechanism of Fe 3+ binding inhibition to Vibrio metschnikovii ferric ion-binding protein, FbpA, by rosmarinic acid and its hydrolysate, danshensu.
Protein Sci., 33, 2024
|
|
8J4J
 
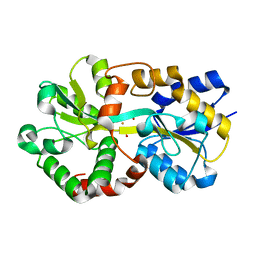 | | X-ray structure of a ferric ion-binding protein A (FbpA) from Vibrio metschnikovii in complex with ferric ion | | Descriptor: | CARBONATE ION, FE (III) ION, Ferric iron ABC transporter iron-binding protein | | Authors: | Lu, P, Jiang, J, Nagata, K. | | Deposit date: | 2023-04-20 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Molecular mechanism of Fe 3+ binding inhibition to Vibrio metschnikovii ferric ion-binding protein, FbpA, by rosmarinic acid and its hydrolysate, danshensu.
Protein Sci., 33, 2024
|
|
3W7Z
 
 | |
6KJL
 
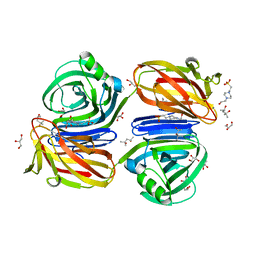 | | Xylanase J from Bacillus sp. strain 41M-1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Manami, S, Teisuke, T, Nakatani, K, Katano, K, Kojima, K, Saka, N, Mikami, B, Yatsunami, R, Nakamura, S, Yasukawa, K. | | Deposit date: | 2019-07-22 | | Release date: | 2019-09-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Increase in the thermostability of GH11 xylanase XynJ from Bacillus sp. strain 41M-1 using site saturation mutagenesis.
Enzyme.Microb.Technol., 130, 2019
|
|
7EHF
 
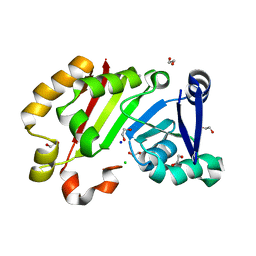 | | Crystal structure of the aminoglycoside resistance methyltransferase NpmB1 | | Descriptor: | 1,2-ETHANEDIOL, 16S rRNA methyltransferase, CHLORIDE ION, ... | | Authors: | Kawai, A, Doi, Y. | | Deposit date: | 2021-03-29 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Functional and Structural Characterization of Acquired 16S rRNA Methyltransferase NpmB1 Conferring Pan-Aminoglycoside Resistance.
Antimicrob.Agents Chemother., 65, 2021
|
|
3B21
 
 | |
7XY8
 
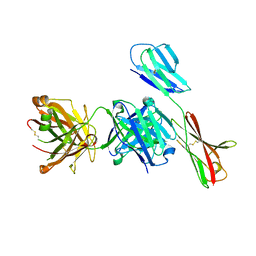 | | Crystal structure of antibody Fab fragment in complex with CD147(EMMPIRIN) | | Descriptor: | Isoform 2 of Basigin, heavy chain, light chain | | Authors: | Nakamura, K, Amano, M, Yoneda, K, Suzuki, M, Fukuchi, K. | | Deposit date: | 2022-06-01 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Novel Antibody Exerts Antitumor Effect through Downregulation of CD147 and Activation of Multiple Stress Signals.
J Oncol, 2022, 2022
|
|
3WC4
 
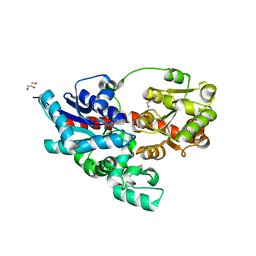 | | Crystal structure of UDP-glucose: anthocyanidin 3-O-glucosyltransferase from Clitoria ternatea | | Descriptor: | ACETATE ION, GLYCEROL, UDP-glucose:anthocyanidin 3-O-glucosyltransferase | | Authors: | Hiromoto, T, Honjo, E, Tamada, T, Kuroki, R. | | Deposit date: | 2013-05-24 | | Release date: | 2013-10-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of UDP-glucose:anthocyanidin 3-O-glucosyltransferase from Clitoria ternatea
J.SYNCHROTRON RADIAT., 20, 2013
|
|
2EBS
 
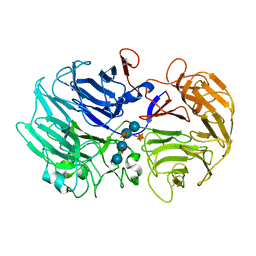 | | Crystal Structure Anaalysis of Oligoxyloglucan reducing-end-specific cellobiohydrolase (OXG-RCBH) D465N Mutant Complexed with a Xyloglucan Heptasaccharide | | Descriptor: | Oligoxyloglucan reducing end-specific cellobiohydrolase, alpha-D-xylopyranose-(1-6)-beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Yaoi, K, Kondo, H, Hiyoshi, A, Noro, N, Sugimoto, H, Miyazaki, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-09 | | Release date: | 2007-06-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Structural Basis for the Exo-mode of Action in GH74 Oligoxyloglucan Reducing End-specific Cellobiohydrolase.
J.Mol.Biol., 370, 2007
|
|
3W3E
 
 | |
