1NOY
 
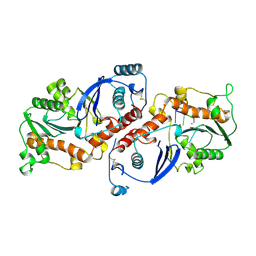 | | DNA POLYMERASE (E.C.2.7.7.7)/DNA COMPLEX | | Descriptor: | DNA (5'-D(*TP*TP*T)-3'), MANGANESE (II) ION, PROTEIN (DNA POLYMERASE (E.C.2.7.7.7)), ... | | Authors: | Wang, J, Yu, P, Lin, T.C, Konigsberg, W.H, Steitz, T.A. | | Deposit date: | 1996-02-16 | | Release date: | 1996-10-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of an NH2-terminal fragment of T4 DNA polymerase and its complexes with single-stranded DNA and with divalent metal ions.
Biochemistry, 35, 1996
|
|
1K8A
 
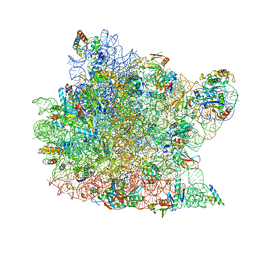 | | Co-crystal structure of Carbomycin A bound to the 50S ribosomal subunit of Haloarcula marismortui | | Descriptor: | 23S RRNA, 5S RRNA, CADMIUM ION, ... | | Authors: | Hansen, J.L, Ippolito, J.A, Ban, N, Nissen, P, Moore, P.B, Steitz, T. | | Deposit date: | 2001-10-23 | | Release date: | 2002-07-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structures of four macrolide antibiotics bound to the large ribosomal subunit.
Mol.Cell, 10, 2002
|
|
4IQJ
 
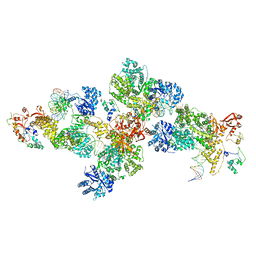 | | Structure of PolIIIalpha-Tauc-DNA complex suggests an atomic model of the replisome | | Descriptor: | DNA (5'-D(*TP*TP*TP*TP*TP*TP*TP*GP*TP*GP*GP*CP*AP*CP*TP*GP*GP*CP*CP*GP*TP*CP*GP*TP*TP*TP*CP*G)-3'), DNA (5'-D(P*CP*GP*AP*AP*AP*CP*GP*AP*CP*GP*GP*CP*CP*AP*GP*TP*GP*CP*CP*A)-3'), DNA (5'-D(P*CP*GP*AP*AP*AP*CP*GP*AP*CP*GP*GP*CP*CP*AP*GP*TP*GP*CP*CP*AP*(DOC))-3'), ... | | Authors: | Liu, B, Lin, J, Steitz, T. | | Deposit date: | 2013-01-11 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of PolIIIalpha-Tauc-DNA complex suggests an atomic model of the replisome
Structure, 21, 2013
|
|
3CC4
 
 | |
1ARO
 
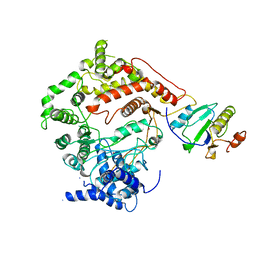 | | T7 RNA POLYMERASE COMPLEXED WITH T7 LYSOZYME | | Descriptor: | MERCURY (II) ION, T7 LYSOZYME, T7 RNA POLYMERASE | | Authors: | Steitz, T, Jeruzalmi, D. | | Deposit date: | 1997-08-08 | | Release date: | 1998-10-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of T7 RNA polymerase complexed to the transcriptional inhibitor T7 lysozyme.
EMBO J., 17, 1998
|
|
3CC2
 
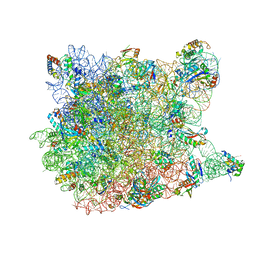 | |
2CHA
 
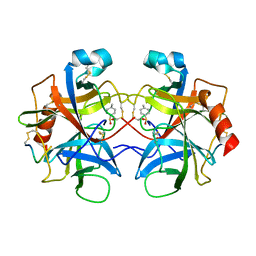 | |
1DPI
 
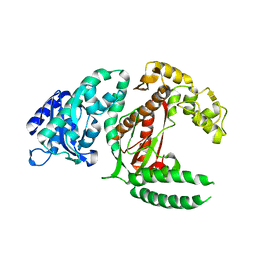 | |
3FWE
 
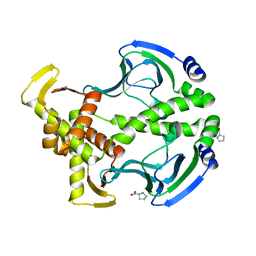 | | Crystal Structure of the Apo D138L CAP mutant | | Descriptor: | Catabolite gene activator, PROLINE | | Authors: | Sharma, H, Wang, J, Kong, J, Yu, S, Steitz, T. | | Deposit date: | 2009-01-17 | | Release date: | 2009-09-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of apo-CAP reveals that large conformational changes are necessary for DNA binding
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4V9F
 
 | |
4V6A
 
 | | Structure of EF-P bound to the 70S ribosome. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Stanley, R.E, Blaha, G. | | Deposit date: | 2009-06-15 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Formation of the first peptide bond: the structure of EF-P bound to the 70S ribosome.
Science, 325, 2009
|
|
4V7L
 
 | | The structures of viomycin bound to the 70S ribosome. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Stanley, R.E, Blaha, G. | | Deposit date: | 2009-11-12 | | Release date: | 2014-07-09 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structures of the anti-tuberculosis antibiotics viomycin and capreomycin bound to the 70S ribosome.
Nat.Struct.Mol.Biol., 17, 2010
|
|
4V7M
 
 | | The structures of Capreomycin bound to the 70S ribosome. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Stanley, R.E, Blaha, G. | | Deposit date: | 2009-11-12 | | Release date: | 2014-07-09 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | The structures of the anti-tuberculosis antibiotics viomycin and capreomycin bound to the 70S ribosome.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3CCU
 
 | |
3CCL
 
 | |
3CCV
 
 | |
3CC7
 
 | |
3CCE
 
 | |
3CCM
 
 | |
3CCS
 
 | |
3CCJ
 
 | |
3CCR
 
 | |
4CHA
 
 | |
1KI3
 
 | | CRYSTAL STRUCTURE OF THYMIDINE KINASE FROM HERPES SIMPLEX VIRUS TYPE I COMPLEXED WITH PENCICLOVIR | | Descriptor: | 9-(4-HYDROXY-3-(HYDROXYMETHYL)BUT-1-YL)GUANINE, SULFATE ION, THYMIDINE KINASE | | Authors: | Champness, J.N, Bennett, M.S, Wien, F, Visse, R, Jarvest, R.L, Summers, W.C, Sanderson, M.R. | | Deposit date: | 1998-05-15 | | Release date: | 1999-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Exploring the active site of herpes simplex virus type-1 thymidine kinase by X-ray crystallography of complexes with aciclovir and other ligands.
Proteins, 32, 1998
|
|
4TQD
 
 | | Crystal Structure of the C-terminal domain of IFRS bound with 3-iodo-L-Phe and ATP | | Descriptor: | 1,2-ETHANEDIOL, 3-iodo-L-phenylalanine, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Nakamura, A, O'Donoghue, P, Soll, D. | | Deposit date: | 2014-06-11 | | Release date: | 2014-11-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1429 Å) | | Cite: | Polyspecific pyrrolysyl-tRNA synthetases from directed evolution.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
