8Z45
 
 | | SFX structure of CraCRY 300 us after photoexcitation of the oxidized protein | | Descriptor: | CHLORIDE ION, Cryptochrome photoreceptor, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Maestre-Reyna, M, Hosokawa, Y, Wang, P.-H, Saft, M, Caramello, N, Engilberge, S, Franz-Badur, S, Ngura Putu, E.P.G, Nakamura, M, Wu, W.-J, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Yang, C.-H, Lin, W.-T, Yang, K.-C, Ban, Y, Imura, T, Kazuoka, A, Tanida, E, Owada, S, Joti, Y, Tanaka, R, Tanaka, T, Luo, F, Tono, K, Kiontke, S, Korf, L, Umena, Y, Tosha, T, Bessho, Y, Nango, E, Iwata, S, Royant, A, Tsai, M.-D, Yamamoto, J, Essen, L.-O. | | Deposit date: | 2024-04-16 | | Release date: | 2025-05-14 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Capturing structural intermediates in an animal-like cryptochrome photoreceptor by time-resolved crystallography.
Sci Adv, 11, 2025
|
|
8Z2D
 
 | | SFX structure of CraCRY 100 ns after photoexcitation of the oxidized protein | | Descriptor: | CHLORIDE ION, Cryptochrome photoreceptor, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Maestre-Reyna, M, Hosokawa, Y, Wang, P.-H, Saft, M, Caramello, N, Engilberge, S, Franz-Badur, S, Ngura Putu, E.P.G, Nakamura, M, Wu, W.-J, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Yang, C.-H, Lin, W.-T, Yang, K.-C, Ban, Y, Imura, T, Kazuoka, A, Tanida, E, Owada, S, Joti, Y, Tanaka, R, Tanaka, T, Luo, F, Tono, K, Kiontke, S, Korf, L, Umena, Y, Tosha, T, Bessho, Y, Nango, E, Iwata, S, Royant, A, Tsai, M.-D, Yamamoto, J, Essen, L.-O. | | Deposit date: | 2024-04-12 | | Release date: | 2025-05-14 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Capturing structural intermediates in an animal-like cryptochrome photoreceptor by time-resolved crystallography.
Sci Adv, 11, 2025
|
|
8Z3D
 
 | | SFX structure of CraCRY 1 us after photoexcitation of the oxidized protein | | Descriptor: | CHLORIDE ION, Cryptochrome photoreceptor, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Maestre-Reyna, M, Hosokawa, Y, Wang, P.-H, Saft, M, Caramello, N, Engilberge, S, Franz-Badur, S, Ngura Putu, E.P.G, Nakamura, M, Wu, W.-J, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Yang, C.-H, Lin, W.-T, Yang, K.-C, Ban, Y, Imura, T, Kazuoka, A, Tanida, E, Owada, S, Joti, Y, Tanaka, R, Tanaka, T, Luo, F, Tono, K, Kiontke, S, Korf, L, Umena, Y, Tosha, T, Bessho, Y, Nango, E, Iwata, S, Royant, A, Tsai, M.-D, Yamamoto, J, Essen, L.-O. | | Deposit date: | 2024-04-15 | | Release date: | 2025-05-14 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Capturing structural intermediates in an animal-like cryptochrome photoreceptor by time-resolved crystallography.
Sci Adv, 11, 2025
|
|
8Z44
 
 | | SFX structure of CraCRY 300 ns after photoexcitation of the oxidized protein | | Descriptor: | CHLORIDE ION, Cryptochrome photoreceptor, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Maestre-Reyna, M, Hosokawa, Y, Wang, P.-H, Saft, M, Caramello, N, Engilberge, S, Franz-Badur, S, Ngura Putu, E.P.G, Nakamura, M, Wu, W.-J, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Yang, C.-H, Lin, W.-T, Yang, K.-C, Ban, Y, Imura, T, Kazuoka, A, Tanida, E, Owada, S, Joti, Y, Tanaka, R, Tanaka, T, Luo, F, Tono, K, Kiontke, S, Korf, L, Umena, Y, Tosha, T, Bessho, Y, Nango, E, Iwata, S, Royant, A, Tsai, M.-D, Yamamoto, J, Essen, L.-O. | | Deposit date: | 2024-04-16 | | Release date: | 2025-05-14 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Capturing structural intermediates in an animal-like cryptochrome photoreceptor by time-resolved crystallography.
Sci Adv, 11, 2025
|
|
8Z6I
 
 | | SFX structure of CraCRY 100 ms after photoexcitation of the oxidized protein | | Descriptor: | CHLORIDE ION, Cryptochrome photoreceptor, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Maestre-Reyna, M, Hosokawa, Y, Wang, P.-H, Saft, M, Caramello, N, Engilberge, S, Franz-Badur, S, Ngura Putu, E.P.G, Nakamura, M, Wu, W.-J, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Yang, C.-H, Lin, W.-T, Yang, K.-C, Ban, Y, Imura, T, Kazuoka, A, Tanida, E, Owada, S, Joti, Y, Tanaka, R, Tanaka, T, Luo, F, Tono, K, Kiontke, S, Korf, L, Umena, Y, Tosha, T, Bessho, Y, Nango, E, Iwata, S, Royant, A, Tsai, M.-D, Yamamoto, J, Essen, L.-O. | | Deposit date: | 2024-04-19 | | Release date: | 2025-05-14 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Capturing structural intermediates in an animal-like cryptochrome photoreceptor by time-resolved crystallography.
Sci Adv, 11, 2025
|
|
8ZA8
 
 | | SFX structure of CraCRY in the fully reduced state | | Descriptor: | CHLORIDE ION, Cryptochrome photoreceptor, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Maestre-Reyna, M, Hosokawa, Y, Wang, P.-H, Saft, M, Caramello, N, Engilberge, S, Franz-Badur, S, Ngura Putu, E.P.G, Nakamura, M, Wu, W.-J, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Yang, C.-H, Lin, W.-T, Yang, K.-C, Ban, Y, Imura, T, Kazuoka, A, Tanida, E, Owada, S, Joti, Y, Tanaka, R, Tanaka, T, Luo, F, Tono, K, Kiontke, S, Korf, L, Umena, Y, Tosha, T, Bessho, Y, Nango, E, Iwata, S, Royant, A, Tsai, M.-D, Yamamoto, J, Essen, L.-O. | | Deposit date: | 2024-04-24 | | Release date: | 2025-05-14 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Capturing structural intermediates in an animal-like cryptochrome photoreceptor by time-resolved crystallography.
Sci Adv, 11, 2025
|
|
8Z6J
 
 | | SFX structure of CraCRY 133 ms after photoexcitation of the oxidized protein | | Descriptor: | CHLORIDE ION, Cryptochrome photoreceptor, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Maestre-Reyna, M, Hosokawa, Y, Wang, P.-H, Saft, M, Caramello, N, Engilberge, S, Franz-Badur, S, Ngura Putu, E.P.G, Nakamura, M, Wu, W.-J, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Yang, C.-H, Lin, W.-T, Yang, K.-C, Ban, Y, Imura, T, Kazuoka, A, Tanida, E, Owada, S, Joti, Y, Tanaka, R, Tanaka, T, Luo, F, Tono, K, Kiontke, S, Korf, L, Umena, Y, Tosha, T, Bessho, Y, Nango, E, Iwata, S, Royant, A, Tsai, M.-D, Yamamoto, J, Essen, L.-O. | | Deposit date: | 2024-04-19 | | Release date: | 2025-05-14 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Capturing structural intermediates in an animal-like cryptochrome photoreceptor by time-resolved crystallography.
Sci Adv, 11, 2025
|
|
8Z3G
 
 | | SFX structure of CraCRY 3 us after photoexcitation of the oxidized protein | | Descriptor: | CHLORIDE ION, Cryptochrome photoreceptor, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Maestre-Reyna, M, Hosokawa, Y, Wang, P.-H, Saft, M, Caramello, N, Engilberge, S, Franz-Badur, S, Ngura Putu, E.P.G, Nakamura, M, Wu, W.-J, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Yang, C.-H, Lin, W.-T, Yang, K.-C, Ban, Y, Imura, T, Kazuoka, A, Tanida, E, Owada, S, Joti, Y, Tanaka, R, Tanaka, T, Luo, F, Tono, K, Kiontke, S, Korf, L, Umena, Y, Tosha, T, Bessho, Y, Nango, E, Iwata, S, Royant, A, Tsai, M.-D, Yamamoto, J, Essen, L.-O. | | Deposit date: | 2024-04-15 | | Release date: | 2025-05-14 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Capturing structural intermediates in an animal-like cryptochrome photoreceptor by time-resolved crystallography.
Sci Adv, 11, 2025
|
|
8Z4M
 
 | | SFX structure of CraCRY 7 ms after photoexcitation of the oxidized protein | | Descriptor: | CHLORIDE ION, Cryptochrome photoreceptor, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Maestre-Reyna, M, Hosokawa, Y, Wang, P.-H, Saft, M, Caramello, N, Engilberge, S, Franz-Badur, S, Ngura Putu, E.P.G, Nakamura, M, Wu, W.-J, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Yang, C.-H, Lin, W.-T, Yang, K.-C, Ban, Y, Imura, T, Kazuoka, A, Tanida, E, Owada, S, Joti, Y, Tanaka, R, Tanaka, T, Luo, F, Tono, K, Kiontke, S, Korf, L, Umena, Y, Tosha, T, Bessho, Y, Nango, E, Iwata, S, Royant, A, Tsai, M.-D, Yamamoto, J, Essen, L.-O. | | Deposit date: | 2024-04-17 | | Release date: | 2025-05-14 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Capturing structural intermediates in an animal-like cryptochrome photoreceptor by time-resolved crystallography.
Sci Adv, 11, 2025
|
|
6QL4
 
 | | Crystal structure of nucleotide-free Mgm1 | | Descriptor: | 1,2-ETHANEDIOL, Putative mitochondrial dynamin protein | | Authors: | Faelber, K, Dietrich, L, Noel, J.K, Wollweber, F, Pfitzner, A.-K, Muehleip, A, Sanchez, R, Kudryashev, M, Chiaruttin, N, Lilie, H, Schlegel, J, Rosenbaum, E, Hessenberger, M, Matthaeus, C, Noe, F, Roux, A, vanderLaan, M, Kuehlbrandt, W, Daumke, O. | | Deposit date: | 2019-01-31 | | Release date: | 2019-07-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure and assembly of the mitochondrial membrane remodelling GTPase Mgm1.
Nature, 571, 2019
|
|
8Y6V
 
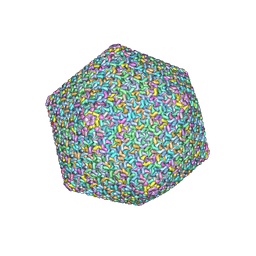 | | Near-atomic structure of icosahedrally averaged jumbo bacteriophage PhiKZ capsid | | Descriptor: | gp119, gp120, gp162, ... | | Authors: | Yang, Y, Shao, Q, Guo, M, Han, L, Zhao, X, Wang, A, Li, X, Wang, B, Pan, J, Chen, Z, Fokine, A, Sun, L, Fang, Q. | | Deposit date: | 2024-02-03 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Capsid structure of bacteriophage Phi KZ provides insights into assembly and stabilization of jumbo phages.
Nat Commun, 15, 2024
|
|
6PVK
 
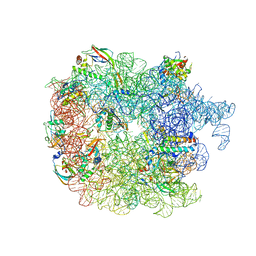 | | Bacterial 45SRbgA ribosomal particle class A | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Ortega, J, Seffouh, A, Jain, N, Jahagirdar, D, Basu, K, Razi, A, Ni, X, Guarne, A, Britton, R.A. | | Deposit date: | 2019-07-20 | | Release date: | 2019-09-18 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural consequences of the interaction of RbgA with a 50S ribosomal subunit assembly intermediate.
Nucleic Acids Res., 47, 2019
|
|
6W1P
 
 | | RT XFEL structure of the one-flash state of Photosystem II (1F, S2-rich) at 2.26 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Ibrahim, M, Fransson, T, Chatterjee, R, Cheah, M.H, Hussein, R, Lassalle, L, Sutherlin, K.D, Young, I.D, Fuller, F.D, Gul, S, Kim, I.-S, Simon, P.S, de Lichtenberg, C, Chernev, P, Bogacz, I, Pham, C, Orville, A.M, Saichek, N, Northen, T.R, Batyuk, A, Carbajo, S, Alonso-Mori, R, Tono, K, Owada, S, Bhowmick, A, Bolotovski, R, Mendez, D, Moriarty, N.W, Holton, J.M, Dobbek, H, Brewster, A.S, Adams, P.D, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2020-03-04 | | Release date: | 2020-05-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Untangling the sequence of events during the S2→ S3transition in photosystem II and implications for the water oxidation mechanism.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6W1R
 
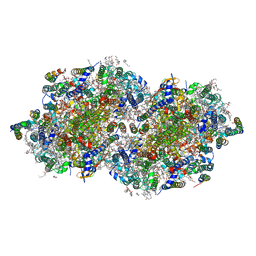 | | RT XFEL structure of Photosystem II 150 microseconds after the second illumination at 2.23 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Ibrahim, M, Fransson, T, Chatterjee, R, Cheah, M.H, Hussein, R, Lassalle, L, Sutherlin, K.D, Young, I.D, Fuller, F.D, Gul, S, Kim, I.-S, Simon, P.S, de Lichtenberg, C, Chernev, P, Bogacz, I, Pham, C, Orville, A.M, Saichek, N, Northen, T.R, Batyuk, A, Carbajo, S, Alonso-Mori, R, Tono, K, Owada, S, Bhowmick, A, Bolotovski, R, Mendez, D, Moriarty, N.W, Holton, J.M, Dobbek, H, Brewster, A.S, Adams, P.D, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2020-03-04 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Untangling the sequence of events during the S2→ S3transition in photosystem II and implications for the water oxidation mechanism.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5WCG
 
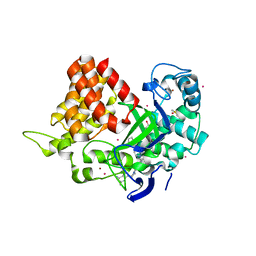 | | SET and MYND Domain Containing protein 2 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DIMETHYL SULFOXIDE, N-lysine methyltransferase SMYD2, ... | | Authors: | Dong, A, Zeng, H, Walker, J.R, Hutch, A, Seitova, A, Tatlock, J, Kumpf, R, Owen, A, Taylor, A, Casimiro-Garcia, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-06-30 | | Release date: | 2017-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The crystal structure of SMYD2 in complex with compound MTF003
to be published
|
|
6WPT
 
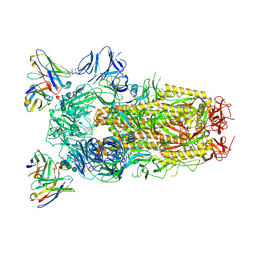 | | Structure of the SARS-CoV-2 spike glycoprotein in complex with the S309 neutralizing antibody Fab fragment (open state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S309 neutralizing antibody heavy chain, ... | | Authors: | Pinto, D, Park, Y.J, Beltramello, M, Walls, A.C, Tortorici, M.A, Bianchi, S, Jaconi, S, Culap, K, Zatta, F, De Marco, A, Peter, A, Guarino, B, Spreafico, R, Cameroni, E, Case, J.B, Chen, R.E, Havenar-Daughton, C, Snell, G, Virgin, H.W, Lanzavecchia, A, Diamond, M.S, Fink, K, Veesler, D, Corti, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2020-04-27 | | Release date: | 2020-05-27 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody.
Nature, 583, 2020
|
|
6W1V
 
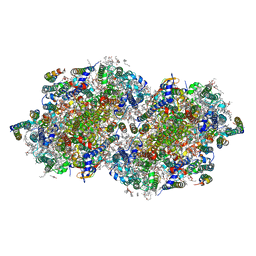 | | RT XFEL structure of the two-flash state of Photosystem II (2F, S3-rich) at 2.09 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Ibrahim, M, Fransson, T, Chatterjee, R, Cheah, M.H, Hussein, R, Lassalle, L, Sutherlin, K.D, Young, I.D, Fuller, F.D, Gul, S, Kim, I.-S, Simon, P.S, de Lichtenberg, C, Chernev, P, Bogacz, I, Pham, C, Orville, A.M, Saichek, N, Northen, T.R, Batyuk, A, Carbajo, S, Alonso-Mori, R, Tono, K, Owada, S, Bhowmick, A, Bolotovski, R, Mendez, D, Moriarty, N.W, Holton, J.M, Dobbek, H, Brewster, A.S, Adams, P.D, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2020-03-04 | | Release date: | 2020-06-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Untangling the sequence of events during the S2→ S3transition in photosystem II and implications for the water oxidation mechanism.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4Y55
 
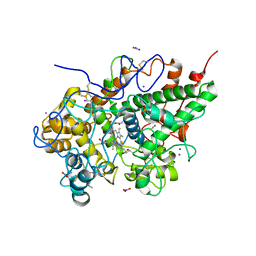 | | Crystal structure of Buffalo lactoperoxidase with Rhodanide at 2.09 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, IODIDE ION, ... | | Authors: | Gupta, A, Tyagi, T.K, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2015-02-11 | | Release date: | 2015-03-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of Buffalo lactoperoxidase with Rhodanide at 2.09 Angstrom resolution
To Be Published
|
|
8G8C
 
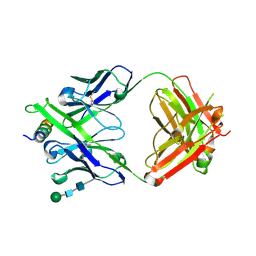 | | Crystal structure of DH1322.1 Fab in complex with HIV proximal MPER peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DH1322.1 heavy chain, DH1322.1 light chain, ... | | Authors: | Niyongabo, A, Janus, B.M, Ofek, G. | | Deposit date: | 2023-02-17 | | Release date: | 2024-05-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Vaccine induction of heterologous HIV-1-neutralizing antibody B cell lineages in humans.
Cell, 187, 2024
|
|
8OOM
 
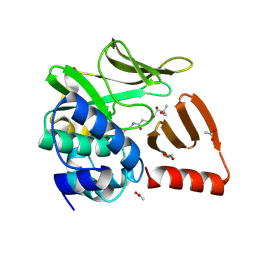 | | Structural and functional studies of geldanamycin amide synthase ShGdmF | | Descriptor: | (2R)-2,4-dihydroxy-3,3-dimethyl-N-{3-oxo-3-[(2-sulfanylethyl)amino]propyl}butanamide, ACETATE ION, GdmF | | Authors: | Ewert, W, Zeilinger, C, Kirschning, A, Preller, M. | | Deposit date: | 2023-04-05 | | Release date: | 2024-04-17 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure and function of the geldanamycin amide synthase from Streptomyces hygroscopicus.
Nat Commun, 16, 2025
|
|
6W1O
 
 | | RT XFEL structure of the dark-stable state of Photosystem II (0F, S1-rich) at 2.08 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Ibrahim, M, Fransson, T, Chatterjee, R, Cheah, M.H, Hussein, R, Lassalle, L, Sutherlin, K.D, Young, I.D, Fuller, F.D, Gul, S, Kim, I.-S, Simon, P.S, de Lichtenberg, C, Chernev, P, Bogacz, I, Pham, C, Orville, A.M, Saichek, N, Northen, T.R, Batyuk, A, Carbajo, S, Alonso-Mori, R, Tono, K, Owada, S, Bhowmick, A, Bolotovski, R, Mendez, D, Moriarty, N.W, Holton, J.M, Dobbek, H, Brewster, A.S, Adams, P.D, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2020-03-04 | | Release date: | 2020-05-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Untangling the sequence of events during the S2→ S3transition in photosystem II and implications for the water oxidation mechanism.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8OSZ
 
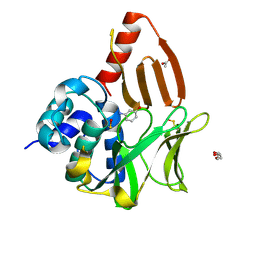 | | Structural and functional studies of geldanamycin amide synthase ShGdmF | | Descriptor: | 3-aminophenol, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ewert, W, Zeilinger, C, Kirschning, A, Preller, M. | | Deposit date: | 2023-04-20 | | Release date: | 2024-05-01 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Structure and function of the geldanamycin amide synthase from Streptomyces hygroscopicus.
Nat Commun, 16, 2025
|
|
8OSV
 
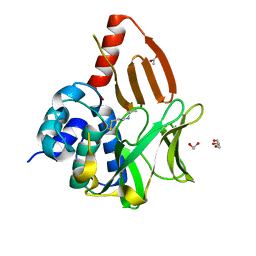 | | Structural and functional studies of geldanamycin amide synthase ShGdmF | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ewert, W, Zeilinger, C, Kirschning, A, Preller, M. | | Deposit date: | 2023-04-20 | | Release date: | 2024-05-01 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Structure and function of the geldanamycin amide synthase from Streptomyces hygroscopicus.
Nat Commun, 16, 2025
|
|
8OT2
 
 | | Structural and functional studies of geldanamycin amide synthase ShGdmF | | Descriptor: | 1,2-ETHANEDIOL, 3-azanyl-5-methyl-phenol, ACETATE ION, ... | | Authors: | Ewert, W, Zeilinger, C, Kirschning, A, Preller, M. | | Deposit date: | 2023-04-20 | | Release date: | 2024-05-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural and functional studies of geldanamycin amide synthase ShGdmF
To Be Published
|
|
6W1Q
 
 | | RT XFEL structure of Photosystem II 50 microseconds after the second illumination at 2.27 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Ibrahim, M, Fransson, T, Chatterjee, R, Cheah, M.H, Hussein, R, Lassalle, L, Sutherlin, K.D, Young, I.D, Fuller, F.D, Gul, S, Kim, I.-S, Simon, P.S, de Lichtenberg, C, Chernev, P, Bogacz, I, Pham, C, Orville, A.M, Saichek, N, Northen, T.R, Batyuk, A, Carbajo, S, Alonso-Mori, R, Tono, K, Owada, S, Bhowmick, A, Bolotovski, R, Mendez, D, Moriarty, N.W, Holton, J.M, Dobbek, H, Brewster, A.S, Adams, P.D, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2020-03-04 | | Release date: | 2020-05-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Untangling the sequence of events during the S2→ S3transition in photosystem II and implications for the water oxidation mechanism.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
