5OB6
 
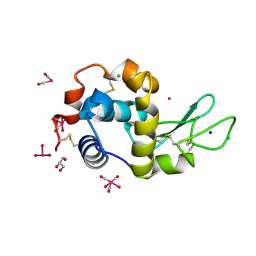 | | X-ray structure of the adduct formed upon reaction of lysozyme with the compound fac-[RuII(CO)3Cl2(N3-IM), IM=imidazole | | Descriptor: | CHLORIDE ION, GLYCEROL, Lysozyme C, ... | | Authors: | Pontillo, N, Ferraro, G, Merlino, A. | | Deposit date: | 2017-06-26 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Ru-Based CO releasing molecules with azole ligands: interaction with proteins and the CO release mechanism disclosed by X-ray crystallography.
Dalton Trans, 46, 2017
|
|
5OBD
 
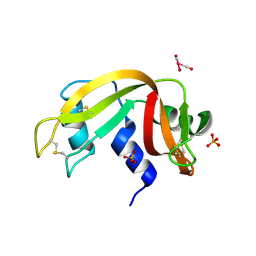 | | X-ray structure of the adduct formed upon reaction of ribonuclease A with the compound fac-[RuII(CO)3Cl2(N3-MIM), MIM=methyl-imidazole | | Descriptor: | PHOSPHATE ION, Ribonuclease pancreatic, tris(oxidaniumylidynemethyl)-tris(oxidanyl)ruthenium | | Authors: | Pontillo, N, Ferraro, G, Merlino, A. | | Deposit date: | 2017-06-26 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Ru-Based CO releasing molecules with azole ligands: interaction with proteins and the CO release mechanism disclosed by X-ray crystallography.
Dalton Trans, 46, 2017
|
|
5OB8
 
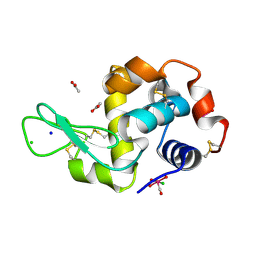 | | X-ray structure of the adduct formed upon reaction of lysozyme with the compound fac-[RuII(CO)3Cl2(N3-MIM), MIM=methyl-imidazole (crystals grown using NaCl) | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme C, ... | | Authors: | Pontillo, N, Ferraro, G, Merlino, A. | | Deposit date: | 2017-06-26 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Ru-Based CO releasing molecules with azole ligands: interaction with proteins and the CO release mechanism disclosed by X-ray crystallography.
Dalton Trans, 46, 2017
|
|
5N26
 
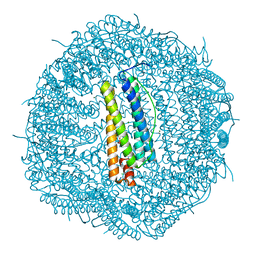 | |
5N27
 
 | |
2Y41
 
 | | Structure of Isopropylmalate dehydrogenase from Thermus thermophilus - complex with IPM and MN | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE, 3-ISOPROPYLMALIC ACID, MANGANESE (II) ION | | Authors: | Graczer, E, merlin, A, Singh, R.K, Manikandan, K, Zavodsky, P, Weiss, M.S, Vas, M. | | Deposit date: | 2011-01-04 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Atomic Level Description of the Domain Closure in a Dimeric Enzyme: Thermus Thermophilus 3-Isopropylmalate Dehydrogenase.
Mol.Biosyst., 7, 2011
|
|
2Y40
 
 | | Structure of Isopropylmalate dehydrogenase from Thermus thermophilus - complex with Mn | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE, MANGANESE (II) ION | | Authors: | Graczer, E, merlin, A, Singh, R.K, Manikandan, K, Zavodsky, P, Weiss, M.S, Vas, M. | | Deposit date: | 2011-01-04 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Atomic Level Description of the Domain Closure in a Dimeric Enzyme: Thermus Thermophilus 3-Isopropylmalate Dehydrogenase.
Mol.Biosyst., 7, 2011
|
|
2Y42
 
 | | Structure of Isopropylmalate dehydrogenase from Thermus thermophilus - complex with NADH and Mn | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE, BICINE, MANGANESE (II) ION, ... | | Authors: | Graczer, E, merlin, A, Singh, R.K, Manikandan, K, Zavodsky, P, Weiss, M.S, Vas, M. | | Deposit date: | 2011-01-04 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Atomic Level Description of the Domain Closure in a Dimeric Enzyme: Thermus Thermophilus 3-Isopropylmalate Dehydrogenase.
Mol.Biosyst., 7, 2011
|
|
2Y3Z
 
 | | Structure of Isopropylmalate dehydrogenase from Thermus thermophilus - apo enzyme | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-ISOPROPYLMALATE DEHYDROGENASE, GLYCEROL, ... | | Authors: | Graczer, E, merlin, A, Singh, R.K, Manikandan, K, Zavodsky, P, Weiss, M.S, Vas, M. | | Deposit date: | 2011-01-04 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Atomic Level Description of the Domain Closure in a Dimeric Enzyme: Thermus Thermophilus 3-Isopropylmalate Dehydrogenase.
Mol.Biosyst., 7, 2011
|
|
1Y92
 
 | | Crystal structure of the P19A/N67D Variant Of Bovine seminal Ribonuclease | | Descriptor: | Seminal ribonuclease | | Authors: | Picone, D, Di Fiore, A, Ercole, C, Franzese, M, Sica, F, Tomaselli, S, Mazzarella, L. | | Deposit date: | 2004-12-14 | | Release date: | 2004-12-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Role of the Hinge Loop in Domain Swapping: THE SPECIAL CASE OF BOVINE SEMINAL RIBONUCLEASE.
J.Biol.Chem., 280, 2005
|
|
1Y94
 
 | | Crystal structure of the G16S/N17T/P19A/S20A/N67D Variant Of Bovine seminal Ribonuclease | | Descriptor: | Seminal ribonuclease | | Authors: | Picone, D, Di Fiore, A, Ercole, C, Franzese, M, Sica, F, Tomaselli, S, Mazzarella, L. | | Deposit date: | 2004-12-14 | | Release date: | 2004-12-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Role of the Hinge Loop in Domain Swapping: THE SPECIAL CASE OF BOVINE SEMINAL RIBONUCLEASE.
J.Biol.Chem., 280, 2005
|
|
5EW1
 
 | | Human thrombin sandwiched between two DNA aptamers: HD22 and HD1-deltaT3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, HD1-deltaT3, ... | | Authors: | Pica, A, Russo Krauss, I, Parente, V, Sica, F. | | Deposit date: | 2015-11-20 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Through-bond effects in the ternary complexes of thrombin sandwiched by two DNA aptamers.
Nucleic Acids Res., 45, 2017
|
|
5EW2
 
 | | Human thrombin sandwiched between two DNA aptamers: HD22 and HD1-deltaT12 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, HD1-deltaT12, ... | | Authors: | Pica, A, Russo Krauss, I, Parente, V, Sica, F. | | Deposit date: | 2015-11-20 | | Release date: | 2016-11-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Through-bond effects in the ternary complexes of thrombin sandwiched by two DNA aptamers.
Nucleic Acids Res., 45, 2017
|
|
6ZCI
 
 | | Crystal structure of BRD4-BD1 in complex with NVS-BET-1 | | Descriptor: | (4~{R})-4-(4-chlorophenyl)-1-cyclopropyl-5-(1,5-dimethyl-6-oxidanylidene-pyridin-3-yl)-3-methyl-4~{H}-pyrrolo[3,4-c]pyrazol-6-one, Bromodomain-containing protein 4 | | Authors: | Faller, M. | | Deposit date: | 2020-06-11 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.976 Å) | | Cite: | BET bromodomain inhibitors regulate keratinocyte plasticity.
Nat.Chem.Biol., 17, 2021
|
|
6TFP
 
 | | BTK in complex with LOU064, a potent and highly selective covalent inhibitor | | Descriptor: | SODIUM ION, Tyrosine-protein kinase BTK, ~{N}-[3-[6-azanyl-5-[2-[methyl(propanoyl)amino]ethoxy]pyrimidin-4-yl]-5-fluoranyl-2-methyl-phenyl]-4-cyclopropyl-2-fluoranyl-benzamide | | Authors: | Scheufler, C, Hinniger, A, Gutmann, S. | | Deposit date: | 2019-11-14 | | Release date: | 2020-03-04 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of LOU064 (Remibrutinib), a Potent and Highly Selective Covalent Inhibitor of Bruton's Tyrosine Kinase.
J.Med.Chem., 63, 2020
|
|
7BFL
 
 | | X-ray structure of SS-RNase-2 des116-120 | | Descriptor: | Angiogenin-1 | | Authors: | Sica, F, Russo Krauss, I, Troisi, R. | | Deposit date: | 2021-01-04 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | The structural features of an ancient ribonuclease from Salmo salar reveal an intriguing case of auto-inhibition.
Int.J.Biol.Macromol., 182, 2021
|
|
7BFK
 
 | | X-ray structure of SS-RNase-2 | | Descriptor: | Angiogenin-1 | | Authors: | Sica, F, Russo Krauss, I, Troisi, R. | | Deposit date: | 2021-01-04 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | The structural features of an ancient ribonuclease from Salmo salar reveal an intriguing case of auto-inhibition.
Int.J.Biol.Macromol., 182, 2021
|
|
6S90
 
 | | BTK in complex with an inhibitor | | Descriptor: | 4-~{tert}-butyl-~{N}-[2-methyl-3-[6-[4-(4-methylpiperazin-1-yl)carbonylphenyl]-7~{H}-pyrrolo[2,3-d]pyrimidin-4-yl]phenyl]benzamide, Tyrosine-protein kinase BTK | | Authors: | Gutmann, S, Hinniger, A. | | Deposit date: | 2019-07-11 | | Release date: | 2019-09-18 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Design of Potent and Selective Covalent Inhibitors of Bruton's Tyrosine Kinase Targeting an Inactive Conformation.
Acs Med.Chem.Lett., 10, 2019
|
|
3TT0
 
 | | Co-structure of Fibroblast Growth Factor Receptor 1 kinase domain with 3-(2,6-dichloro-3,5-dimethoxy-phenyl)-1-{6-[4-(4-ethyl-piperazin-1-yl)-phenylamino]-pyrimidin-4-yl}-1-methyl-urea (BGJ398) | | Descriptor: | 3-(2,6-dichloro-3,5-dimethoxyphenyl)-1-(6-{[4-(4-ethylpiperazin-1-yl)phenyl]amino}pyrimidin-4-yl)-1-methylurea, Basic fibroblast growth factor receptor 1, GLYCEROL, ... | | Authors: | Bussiere, D.E, Murray, J.M, Shu, W. | | Deposit date: | 2011-09-13 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of 3-(2,6-dichloro-3,5-dimethoxy-phenyl)-1-{6-[4-(4-ethyl-piperazin-1-yl)-phenylamino]-pyrimidin-4-yl}-1-methyl-urea (NVP-BGJ398), a potent and selective inhibitor of the fibroblast growth factor receptor family of receptor tyrosine kinase.
J.Med.Chem., 54, 2011
|
|
6ETO
 
 | | Atomic resolution structure of RNase A (data collection 5) | | Descriptor: | ISOPROPYL ALCOHOL, Ribonuclease pancreatic | | Authors: | Caterino, M, Vergara, A, Merlino, A. | | Deposit date: | 2017-10-27 | | Release date: | 2018-02-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | The Alkylquinolone Repertoire of Pseudomonas aeruginosa is Linked to Structural Flexibility of the FabH-like 2-Heptyl-3-hydroxy-4(1H)-quinolone (PQS) Biosynthesis Enzyme PqsBC.
Chembiochem, 2018
|
|
