6KMV
 
 | |
6KN1
 
 | | P20/P12 of caspase-11 mutant C254A | | Descriptor: | Caspase-4 | | Authors: | Ding, J, Sun, Q. | | Deposit date: | 2019-08-02 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Mechanism for GSDMD Targeting by Autoprocessed Caspases in Pyroptosis.
Cell, 180, 2020
|
|
6KN0
 
 | |
6LAT
 
 | | The cryo-EM structure of HEV VLP | | Descriptor: | Protein ORF2 | | Authors: | Zheng, Q, He, M, Li, S. | | Deposit date: | 2019-11-13 | | Release date: | 2019-12-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Viral neutralization by antibody-imposed physical disruption.
Proc.Natl.Acad.Sci.USA, 2019
|
|
6IGD
 
 | | Crystal structure of HPV58/33 chimeric L1 pentamer | | Descriptor: | Major capsid protein L1 | | Authors: | Li, Z.H, Song, S, He, M.Z, Gu, Y, Li, S.W. | | Deposit date: | 2018-09-25 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Rational design of a triple-type human papillomavirus vaccine by compromising viral-type specificity.
Nat Commun, 9, 2018
|
|
6IGF
 
 | | Crystal structure of Human Papillomavirus type 52 pentamer | | Descriptor: | Major capsid protein L1 | | Authors: | Li, Z.H, Song, S, He, M.Z, Gu, Y, Li, S.W. | | Deposit date: | 2018-09-25 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.751 Å) | | Cite: | Rational design of a triple-type human papillomavirus vaccine by compromising viral-type specificity.
Nat Commun, 9, 2018
|
|
6IGE
 
 | | Crystal structure of Human Papillomavirus type 33 pentamer | | Descriptor: | Major capsid protein L1 | | Authors: | Li, Z.H, Song, S, He, M.Z, Gu, Y, Li, S.W. | | Deposit date: | 2018-09-25 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Rational design of a triple-type human papillomavirus vaccine by compromising viral-type specificity.
Nat Commun, 9, 2018
|
|
1T0Y
 
 | | Solution Structure of a Ubiquitin-Like Domain from Tubulin-binding Cofactor B | | Descriptor: | tubulin folding cofactor B | | Authors: | Lytle, B.L, Peterson, F.C, Qui, S.H, Luo, M, Volkman, B.F, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-04-13 | | Release date: | 2004-04-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a Ubiquitin-like Domain from Tubulin-binding Cofactor B.
J.Biol.Chem., 279, 2004
|
|
5KU9
 
 | | Crystal structure of MCL1 with compound 1 | | Descriptor: | (3~{S})-3-azanyl-4-(4-bromophenyl)-~{N}-[(3~{S})-1-[2-[[(2~{R})-1-(3,4-dichlorophenyl)-4-(methylamino)-4-oxidanylidene-butan-2-yl]amino]-2-oxidanylidene-ethyl]-2-oxidanylidene-4,5-dihydro-3~{H}-1-benzazepin-3-yl]butanamide, Induced myeloid leukemia cell differentiation protein Mcl-1, SODIUM ION | | Authors: | Ferguson, A.D. | | Deposit date: | 2016-07-13 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure Based Design of Non-Natural Peptidic Macrocyclic Mcl-1 Inhibitors.
ACS Med Chem Lett, 8, 2017
|
|
6IGC
 
 | | Crystal structure of HPV58/33/52 chimeric L1 pentamer | | Descriptor: | Major capsid protein L1 | | Authors: | Li, Z.H, Song, S, He, M.Z, Gu, Y, Li, S.W. | | Deposit date: | 2018-09-25 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Rational design of a triple-type human papillomavirus vaccine by compromising viral-type specificity.
Nat Commun, 9, 2018
|
|
1Q53
 
 | |
1Q4R
 
 | | Gene Product of At3g17210 from Arabidopsis Thaliana | | Descriptor: | MAGNESIUM ION, protein At3g17210 | | Authors: | Phillips Jr, G.N, Bingman, C.A, Johnson, K.A, Smith, D.W, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2003-08-04 | | Release date: | 2003-11-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the protein from gene At3g17210 of Arabidopsis thaliana
Proteins, 57, 2004
|
|
1Q44
 
 | | Crystal Structure of an Arabidopsis Thaliana Putative Steroid Sulfotransferase | | Descriptor: | MALONIC ACID, Steroid Sulfotransferase | | Authors: | Phillips Jr, G.N, Smith, D.W, Johnson, K.A, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2003-08-01 | | Release date: | 2003-11-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of At2g03760, a putative steroid sulfotransferase from Arabidopsis thaliana
Proteins, 57, 2004
|
|
2Q3M
 
 | | Ensemble refinement of the protein crystal structure of an Arabidopsis thaliana putative steroid sulphotransferase | | Descriptor: | Flavonol sulfotransferase-like, MALONIC ACID | | Authors: | Levin, E.J, Kondrashov, D.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2007-05-30 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ensemble refinement of protein crystal structures: validation and application.
Structure, 15, 2007
|
|
2Q3P
 
 | | Ensemble refinement of the protein crystal structure of At3g17210 from Arabidopsis thaliana | | Descriptor: | MAGNESIUM ION, Uncharacterized protein At3g17210 | | Authors: | Levin, E.J, Kondrashov, D.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2007-05-30 | | Release date: | 2007-06-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ensemble refinement of protein crystal structures: validation and application.
Structure, 15, 2007
|
|
4RAX
 
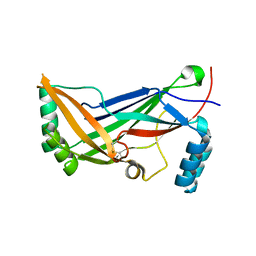 | | A regulatory domain of an ion channel | | Descriptor: | Piezo-type mechanosensitive ion channel component 1 | | Authors: | Ge, J, Yang, M. | | Deposit date: | 2014-09-11 | | Release date: | 2015-09-23 | | Last modified: | 2018-04-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Architecture of the mammalian mechanosensitive Piezo1 channel.
Nature, 527, 2015
|
|
7Y04
 
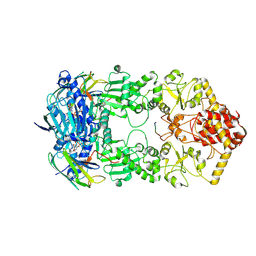 | | Hsp90-AhR-p23 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Aryl hydrocarbon receptor, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Wen, Z.L, Zhai, Y.J, Zhu, Y, Sun, F. | | Deposit date: | 2022-06-03 | | Release date: | 2023-01-04 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of the cytosolic AhR complex.
Structure, 31, 2023
|
|
5CDI
 
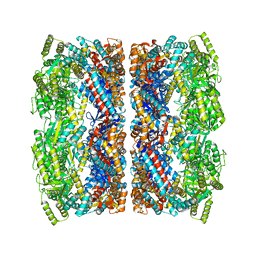 | | Chloroplast chaperonin 60b1 of Chlamydomonas | | Descriptor: | Chaperonin 60B1 | | Authors: | Zhang, S, Zhou, H, Yu, F, Gao, F, He, J, Liu, C. | | Deposit date: | 2015-07-04 | | Release date: | 2016-05-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.807 Å) | | Cite: | Structural insight into the cooperation of chloroplast chaperonin subunits
Bmc Biol., 14, 2016
|
|
5CMA
 
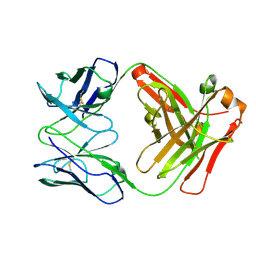 | | Anti-B7-H3 monoclonal antibody ch8H9 Fab fragment | | Descriptor: | Antibody ch8H9 Fab heavy chain, Antibody ch8H9 Fab light chain | | Authors: | Ahmed, M, Goldgur, Y, Cheng, M, Cheung, N.K. | | Deposit date: | 2015-07-16 | | Release date: | 2015-10-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Humanized Affinity-matured Monoclonal Antibody 8H9 Has Potent Antitumor Activity and Binds to FG Loop of Tumor Antigen B7-H3.
J.Biol.Chem., 290, 2015
|
|
6KOX
 
 | |
1DXO
 
 | | Crystal structure of human NAD[P]H-QUINONE oxidoreductase CO with 2,3,5,6,tetramethyl-P-benzoquinone (duroquinone) at 2.5 Angstrom resolution | | Descriptor: | DUROQUINONE, FLAVIN-ADENINE DINUCLEOTIDE, QUINONE REDUCTASE | | Authors: | Faig, M, Bianchet, M.A, Chen, S, Winski, S, Ross, D, Amzel, L.M. | | Deposit date: | 2000-01-12 | | Release date: | 2000-04-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of Recombinant Mouse and Human Nad(P)H:Quinone Oxidoreductases:Species Comparison and Structural Changes with Substrate Binding and Release
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1DXQ
 
 | | CRYSTAL STRUCTURE OF MOUSE NAD[P]H-QUINONE OXIDOREDUCTASE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, QUINONE REDUCTASE | | Authors: | Faig, M, Bianchet, M.A, Chen, S, Winski, S, Ross, D, Amzel, L.M. | | Deposit date: | 2000-01-14 | | Release date: | 2000-04-17 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of Recombinant Mouse and Human Nad(P)H:Quinone Oxidoreductases:Species Comparison and Structural Changes with Substrate Binding and Release
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1D4A
 
 | | CRYSTAL STRUCTURE OF HUMAN NAD[P]H-QUINONE OXIDOREDUCTASE AT 1.7 A RESOLUTION | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, QUINONE REDUCTASE | | Authors: | Faig, M, Bianchet, M.A, Chen, S, Winski, S, Ross, D, Amzel, L.M. | | Deposit date: | 1999-10-01 | | Release date: | 1999-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of recombinant human and mouse NAD(P)H:quinone oxidoreductases: species comparison and structural changes with substrate binding and release.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1GG5
 
 | | CRYSTAL STRUCTURE OF A COMPLEX OF HUMAN NAD[P]H-QUINONE OXIDOREDUCTASE AND A CHEMOTHERAPEUTIC DRUG (E09) AT 2.5 A RESOLUTION | | Descriptor: | 3-HYDROXYMETHYL-5-AZIRIDINYL-1METHYL-2-[1H-INDOLE-4,7-DIONE]-PROPANOL, FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H DEHYDROGENASE [QUINONE] 1 | | Authors: | Faig, M, Bianchet, M.A, Winski, S, Hargreaves, R, Moody, C.J, Hudnott, A.R, Ross, D, Amzel, L.M. | | Deposit date: | 2000-07-12 | | Release date: | 2001-09-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-based development of anticancer drugs: complexes of NAD(P)H:quinone oxidoreductase 1 with chemotherapeutic quinones.
Structure, 9, 2001
|
|
2ETT
 
 | | Solution Structure of Human Sorting Nexin 22 PX Domain | | Descriptor: | Sorting nexin-22 | | Authors: | Song, J, Zhao, Q, Tyler, R.C, Lee, M.S, Newman, C.L, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-10-27 | | Release date: | 2005-11-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human sorting nexin 22.
Protein Sci., 16, 2007
|
|
