7CO7
 
 | | HtrA-type protease AlgWS227A with decapeptide | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, AlgW protein, decapeptide SVRDELRWVF | | Authors: | Li, T, Song, Y.J, Bao, R. | | Deposit date: | 2020-08-04 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular Basis of the Versatile Regulatory Mechanism of HtrA-Type Protease AlgW from Pseudomonas aeruginosa.
Mbio, 12, 2021
|
|
7BDN
 
 | | Structure of the Streptomyces coelicolor small laccase - cubic crystal form | | Descriptor: | COPPER (II) ION, Putative copper oxidase | | Authors: | Zovo, K, Majumdar, S, Lukk, T. | | Deposit date: | 2020-12-22 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Substitution of the Methionine Axial Ligand of the T1 Copper for the Fungal-like Phenylalanine Ligand (M298F) Causes Local Structural Perturbations that Lead to Thermal Instability and Reduced Catalytic Efficiency of the Small Laccase from Streptomyces coelicolor A3(2).
Acs Omega, 7, 2022
|
|
7BFM
 
 | |
7CSV
 
 | | Pseudomonas aeruginosa antitoxin HigA | | Descriptor: | HTH cro/C1-type domain-containing protein | | Authors: | Song, Y.J, Luo, G.H, Bao, R. | | Deposit date: | 2020-08-17 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Pseudomonas aeruginosa antitoxin HigA functions as a diverse regulatory factor by recognizing specific pseudopalindromic DNA motifs.
Environ.Microbiol., 23, 2021
|
|
7WA9
 
 | |
7U3W
 
 | | [L224] Self-assembling tensegrity triangle with two turns, two turns and four turns of DNA per axis by linker addition with P1 symmetry | | Descriptor: | DNA (5'-D(*AP*AP*CP*CP*TP*AP*CP*CP*TP*GP*GP*CP*AP*GP*GP*AP*CP*GP*AP*CP*T)-3'), DNA (5'-D(*AP*GP*AP*GP*TP*CP*GP*TP*GP*GP*CP*TP*CP*G)-3'), DNA (5'-D(*CP*AP*CP*GP*AP*GP*CP*CP*TP*GP*AP*TP*CP*GP*GP*AP*CP*AP*AP*GP*A)-3'), ... | | Authors: | Woloszyn, K, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2022-02-28 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (6.33 Å) | | Cite: | Augmented DNA Nanoarchitectures: A Structural Library of 3D Self-Assembling Tensegrity Triangle Variants.
Adv Mater, 34, 2022
|
|
7U3Z
 
 | | [F244] Self-assembling tensegrity triangle with two turns, four turns and four turns of DNA per axis by extension with P1 symmetry | | Descriptor: | DNA (35-MER), DNA (42-MER), DNA (5'-D(*AP*AP*CP*CP*TP*AP*CP*CP*TP*GP*GP*CP*AP*GP*GP*AP*CP*GP*AP*CP*T)-3'), ... | | Authors: | Woloszyn, K, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2022-02-28 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (7.55 Å) | | Cite: | Augmented DNA Nanoarchitectures: A Structural Library of 3D Self-Assembling Tensegrity Triangle Variants.
Adv Mater, 34, 2022
|
|
7U3V
 
 | | [F224] Self-assembling tensegrity triangle with two turns, two turns and four turns of DNA per axis by extension with P1 symmetry | | Descriptor: | DNA (35-MER), DNA (42-MER), DNA (5'-D(*CP*AP*CP*GP*AP*GP*CP*CP*TP*GP*AP*TP*CP*GP*GP*AP*CP*AP*AP*GP*A)-3'), ... | | Authors: | Woloszyn, K, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2022-02-28 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (5.14 Å) | | Cite: | Augmented DNA Nanoarchitectures: A Structural Library of 3D Self-Assembling Tensegrity Triangle Variants.
Adv Mater, 34, 2022
|
|
7U41
 
 | | [F234] Self-assembling tensegrity triangle with two turns, three turns and four turns of DNA per axis by extension with P1 symmetry | | Descriptor: | DNA (31-MER), DNA (35-MER), DNA (42-MER), ... | | Authors: | Woloszyn, K, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2022-02-28 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (7.24 Å) | | Cite: | Augmented DNA Nanoarchitectures: A Structural Library of 3D Self-Assembling Tensegrity Triangle Variants.
Adv Mater, 34, 2022
|
|
7U42
 
 | | [F334] Self-assembling tensegrity triangle with three turns, three turns and four turns of DNA per axis by extension with P1 symmetry | | Descriptor: | DNA (31-MER), DNA (35-MER), DNA (42-MER), ... | | Authors: | Woloszyn, K, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2022-02-28 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (7.71 Å) | | Cite: | Augmented DNA Nanoarchitectures: A Structural Library of 3D Self-Assembling Tensegrity Triangle Variants.
Adv Mater, 34, 2022
|
|
7U43
 
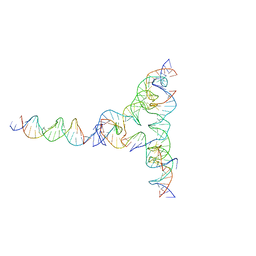 | | [L334] Self-assembling tensegrity triangle with three turns, three turns and four turns of DNA per axis by extension and linker addition with P1 symmetry | | Descriptor: | DNA (31-MER), DNA (5'-D(*TP*AP*CP*AP*CP*CP*GP*AP*TP*CP*AP*CP*CP*TP*GP*CP*CP*AP*CP*CP*G)-3'), DNA (5'-D(P*AP*CP*TP*GP*AP*TP*GP*TP*GP*GP*TP*AP*GP*G)-3'), ... | | Authors: | Woloszyn, K, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2022-02-28 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (7.55 Å) | | Cite: | Augmented DNA Nanoarchitectures: A Structural Library of 3D Self-Assembling Tensegrity Triangle Variants.
Adv Mater, 34, 2022
|
|
7U45
 
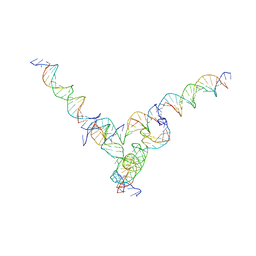 | | [L344] Self-assembling tensegrity triangle with three turns, four turns and four turns of DNA per axis by extension and linker addition with P1 symmetry | | Descriptor: | DNA (31-MER), DNA (5'-D(P*AP*CP*TP*GP*AP*TP*GP*TP*GP*GP*TP*AP*GP*G)-3'), DNA (5'-D(P*AP*GP*GP*CP*AP*GP*CP*CP*TP*GP*TP*AP*CP*GP*GP*AP*CP*AP*TP*CP*A)-3'), ... | | Authors: | Woloszyn, K, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2022-02-28 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (8.05 Å) | | Cite: | Augmented DNA Nanoarchitectures: A Structural Library of 3D Self-Assembling Tensegrity Triangle Variants.
Adv Mater, 34, 2022
|
|
7U3P
 
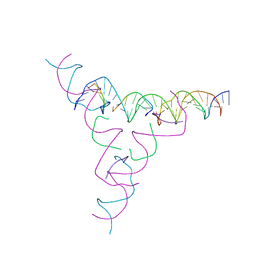 | | [3T7] Self-assembling tensegrity triangle with three turns of DNA per axis with R3 symmetry | | Descriptor: | DNA (31-MER), DNA (5'-D(P*CP*CP*GP*TP*AP*CP*A)-3'), DNA (5'-D(P*GP*GP*CP*TP*GP*C)-3'), ... | | Authors: | Woloszyn, K, Vecchioni, S, Lu, B, Ma, Y, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2022-02-28 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (6.06 Å) | | Cite: | Augmented DNA Nanoarchitectures: A Structural Library of 3D Self-Assembling Tensegrity Triangle Variants.
Adv Mater, 34, 2022
|
|
7U3S
 
 | | [2T7+21] Self-assembling tensegrity triangle with two turns of DNA and the sticky end addition of a two-turn linker per axis with R3 symmetry | | Descriptor: | DNA (5'-D(P*CP*CP*GP*TP*AP*CP*A)-3'), DNA (5'-D(P*GP*AP*GP*CP*AP*GP*CP*CP*TP*GP*TP*AP*CP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(P*GP*AP*TP*GP*CP*TP*GP*AP*CP*GP*TP*AP*GP*TP*AP*GP*CP*AP*GP*AP*G)-3'), ... | | Authors: | Woloszyn, K, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2022-02-28 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (9.5 Å) | | Cite: | Augmented DNA Nanoarchitectures: A Structural Library of 3D Self-Assembling Tensegrity Triangle Variants.
Adv Mater, 34, 2022
|
|
7U3T
 
 | | [F223] Self-assembling tensegrity triangle with two turns, two turns and three turns of DNA per axis by extension with P1 symmetry | | Descriptor: | DNA (31-MER), DNA (5'-D(*AP*AP*CP*CP*TP*AP*CP*CP*TP*GP*GP*CP*AP*GP*GP*AP*CP*GP*AP*CP*T)-3'), DNA (5'-D(*CP*AP*CP*GP*AP*GP*CP*CP*TP*GP*AP*TP*CP*GP*GP*AP*CP*AP*AP*GP*A)-3'), ... | | Authors: | Woloszyn, K, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P, Zhu, E. | | Deposit date: | 2022-02-28 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (4.69 Å) | | Cite: | Augmented DNA Nanoarchitectures: A Structural Library of 3D Self-Assembling Tensegrity Triangle Variants.
Adv Mater, 34, 2022
|
|
7U3R
 
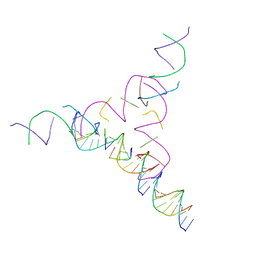 | | [2T7+10] Self-assembling tensegrity triangle with two turns of DNA and the sticky end attachment of a one-turn linker per axis, with R3 symmetry | | Descriptor: | DNA (5'-D(*AP*CP*GP*CP*AP*GP*CP*CP*TP*GP*TP*AP*CP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(*GP*AP*TP*GP*CP*TP*GP*AP*GP*T)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*TP*GP*T)-3'), ... | | Authors: | Woloszyn, K, Vecchioni, S, Lu, B, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2022-02-28 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (6.27 Å) | | Cite: | Augmented DNA Nanoarchitectures: A Structural Library of 3D Self-Assembling Tensegrity Triangle Variants.
Adv Mater, 34, 2022
|
|
7U3Y
 
 | | [L233] Self-assembling tensegrity triangle with two turns, three turns and three turns of DNA per axis by linker addition with P1 symmetry | | Descriptor: | DNA (5'-D(*AP*AP*CP*CP*TP*AP*CP*CP*TP*GP*GP*CP*AP*GP*GP*AP*CP*GP*AP*CP*T)-3'), DNA (5'-D(*AP*CP*TP*GP*AP*TP*GP*TP*GP*GP*TP*AP*GP*G)-3'), DNA (5'-D(*AP*GP*GP*CP*AP*GP*CP*CP*TP*GP*TP*AP*CP*GP*GP*AP*CP*AP*TP*CP*A)-3'), ... | | Authors: | Woloszyn, K, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2022-02-28 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (6.06 Å) | | Cite: | Augmented DNA Nanoarchitectures: A Structural Library of 3D Self-Assembling Tensegrity Triangle Variants.
Adv Mater, 34, 2022
|
|
7U3O
 
 | | [a2T7] Self-assembling asymmetric tensegrity triangle with P1 symmetry | | Descriptor: | DNA (5'-D(*AP*AP*CP*CP*TP*AP*CP*CP*TP*GP*GP*CP*AP*GP*GP*AP*CP*GP*AP*CP*T)-3'), DNA (5'-D(*CP*AP*CP*GP*AP*GP*CP*CP*TP*GP*AP*TP*CP*GP*GP*AP*CP*AP*AP*GP*A)-3'), DNA (5'-D(*GP*AP*GP*CP*GP*AP*CP*CP*TP*GP*TP*AP*CP*GP*GP*AP*CP*AP*TP*CP*A)-3'), ... | | Authors: | Woloszyn, K, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2022-02-28 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Augmented DNA Nanoarchitectures: A Structural Library of 3D Self-Assembling Tensegrity Triangle Variants.
Adv Mater, 34, 2022
|
|
7U3U
 
 | | [L223] Self-assembling tensegrity triangle with two turns, two turns and three turns of DNA per axis by linker addition with P1 symmetry | | Descriptor: | DNA (5'-D(*AP*AP*CP*CP*TP*AP*CP*TP*TP*GP*GP*CP*AP*GP*GP*AP*CP*GP*AP*CP*T)-3'), DNA (5'-D(*AP*CP*GP*CP*GP*AP*CP*CP*TP*GP*TP*AP*CP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(*CP*AP*CP*GP*AP*GP*CP*CP*TP*GP*AP*TP*CP*GP*GP*AP*CP*AP*AP*GP*A)-3'), ... | | Authors: | Woloszyn, K, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2022-02-28 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (4.46 Å) | | Cite: | Augmented DNA Nanoarchitectures: A Structural Library of 3D Self-Assembling Tensegrity Triangle Variants.
Adv Mater, 34, 2022
|
|
7U3X
 
 | | [F233] Self-assembling tensegrity triangle with two turns, three turns and three turns of DNA per axis by extension with P1 symmetry | | Descriptor: | DNA (31-MER), DNA (5'-D(*AP*AP*CP*CP*TP*AP*CP*CP*TP*GP*GP*CP*AP*GP*GP*AP*CP*GP*AP*CP*T)-3'), DNA (5'-D(*TP*CP*TP*AP*GP*CP*AP*TP*AP*GP*AP*CP*TP*GP*AP*TP*GP*TP*GP*GP*TP*AP*GP*G)-3'), ... | | Authors: | Woloszyn, K, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2022-02-28 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (5.68 Å) | | Cite: | Augmented DNA Nanoarchitectures: A Structural Library of 3D Self-Assembling Tensegrity Triangle Variants.
Adv Mater, 34, 2022
|
|
7U40
 
 | | [L244] Self-assembling tensegrity triangle with two turns, four turns and four turns of DNA per axis by linker addition with P1 symmetry | | Descriptor: | DNA (5'-D(*AP*AP*CP*CP*TP*AP*CP*CP*TP*GP*GP*CP*AP*GP*GP*AP*CP*GP*AP*CP*T)-3'), DNA (5'-D(*TP*TP*AP*GP*TP*CP*GP*TP*GP*GP*CP*TP*CP*G)-3'), DNA (5'-D(P*AP*CP*TP*GP*AP*TP*GP*TP*GP*GP*TP*AP*GP*G)-3'), ... | | Authors: | Woloszyn, K, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2022-02-28 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (7.55 Å) | | Cite: | Augmented DNA Nanoarchitectures: A Structural Library of 3D Self-Assembling Tensegrity Triangle Variants.
Adv Mater, 34, 2022
|
|
7U44
 
 | | [F344] Self-assembling tensegrity triangle with three turns, four turns and four turns of DNA per axis by extension with P1 symmetry | | Descriptor: | DNA (31-MER), DNA (35-MER), DNA (42-MER), ... | | Authors: | Woloszyn, K, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2022-02-28 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (8.46 Å) | | Cite: | Augmented DNA Nanoarchitectures: A Structural Library of 3D Self-Assembling Tensegrity Triangle Variants.
Adv Mater, 34, 2022
|
|
5NFN
 
 | |
5NFO
 
 | |
7C83
 
 | | Crystal structure of an integral membrane steroid 5-alpha-reductase PbSRD5A | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3-oxo-5-alpha-steroid 4-dehydrogenase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Ren, R.B, Han, Y.F, Xiao, Q.J, Deng, D. | | Deposit date: | 2020-05-28 | | Release date: | 2021-01-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of steroid reductase SRD5A reveals conserved steroid reduction mechanism.
Nat Commun, 12, 2021
|
|
