8WR3
 
 | | Cryo-EM structure of lysosomal protein LYCHOS | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Wang, Z.H, Qian, H.W. | | Deposit date: | 2023-10-12 | | Release date: | 2025-01-15 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for cholesterol sensing of LYCHOS and its interaction with indoxyl sulfate.
Nat Commun, 16, 2025
|
|
4DVF
 
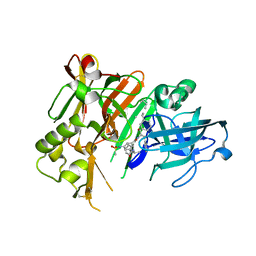 | | Crystal structure of BACE1 with its inhibitor | | Descriptor: | Beta-secretase 1, METHYL (2S)-1-[(2R,5S,8S,12S,13S)-2,13-DIBENZYL-12-HYDROXY-3,5-DIMETHYL-8-(2-METHYLPROPYL)-15-(3-[(METHYLSULFONYL)AMINO]-5-{[(1R)-1-PHENYLETHYL]CARBAMOYL}PHENYL)-4,7,10,15-TETRAOXO-3,6,9,14-TETRAAZAPENTADECAN-1-OYL]PYRROLIDINE-2-CARBOXYLATE | | Authors: | Xu, Y.C, Chen, W.Y, Li, L, Chen, T.T. | | Deposit date: | 2012-02-23 | | Release date: | 2013-01-16 | | Last modified: | 2021-09-15 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Cyanobacterial Peptides as a Prototype for the Design of Potent beta-Secretase Inhibitors and the Development of Selective Chemical Probes for Other Aspartic Proteases
J.Med.Chem., 55, 2012
|
|
9IKY
 
 | | Crystal structure of 1-2C-T96F TCR in complex with HLA-A*11:01 bound to KRAS-G12V peptide (VVGAVGVGK) | | Descriptor: | 1-2C-T96F TCR alpha chain, 1-2C-T96F TCR beta chain, Beta-2-microglobulin, ... | | Authors: | Ma, K.K, Tan, S.G, Gao, G.F, Chai, Y. | | Deposit date: | 2024-06-29 | | Release date: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | An Engineered Soluble Single-Chain TCR Engager for KRAS-G12V Specific Tumor Immunotherapy.
Adv Sci, 2025
|
|
8IJK
 
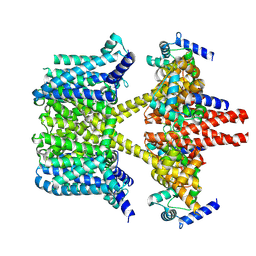 | | human KCNQ2-CaM-Ebio1 complex in the presence of PIP2 | | Descriptor: | Calmodulin-1, N-(1,2-dihydroacenaphthylen-5-yl)-4-fluoranyl-benzamide, Potassium voltage-gated channel subfamily KQT member 2 | | Authors: | Ma, D, Guo, J. | | Deposit date: | 2023-02-27 | | Release date: | 2024-01-17 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | A small-molecule activation mechanism that directly opens the KCNQ2 channel.
Nat.Chem.Biol., 20, 2024
|
|
4DV9
 
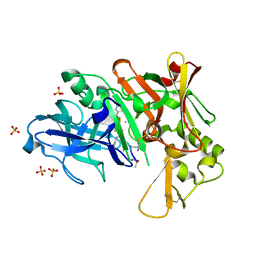 | | Crystal structure of BACE1 with its inhibitor | | Descriptor: | Beta-secretase 1, METHYL (2S)-1-[(2R,5S,8S,12S,13S,16S,19S,22S)-16-(3-AMINO-3-OXOPROPYL)-2,13-DIBENZYL-12,22-DIHYDROXY-3,5,17-TRIMETHYL-8-(2-METHYLPROPYL)-4,7,10,15,18,21-HEXAOXO-19-(PROPAN-2-YL)-3,6,9,14,17,20-HEXAAZATRICOSAN-1-OYL]PYRROLIDINE-2-CARBOXYLATE (NON-PREFERRED NAME), SULFATE ION | | Authors: | Xu, Y.C, Chen, W.Y, Li, L, Chen, T.T. | | Deposit date: | 2012-02-23 | | Release date: | 2013-01-16 | | Last modified: | 2021-09-15 | | Method: | X-RAY DIFFRACTION (2.076 Å) | | Cite: | Cyanobacterial Peptides as a Prototype for the Design of Potent beta-Secretase Inhibitors and the Development of Selective Chemical Probes for Other Aspartic Proteases
J.Med.Chem., 55, 2012
|
|
8JA0
 
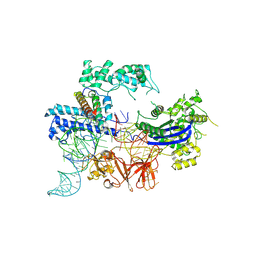 | | Cryo-EM structure of the NmeCas9-sgRNA-AcrIIC4 ternary complex | | Descriptor: | CRISPR-associated endonuclease Cas9, RNA (117-MER), Uncharacterized protein | | Authors: | Yin, H, Li, Z, Yu, G.M, Li, X.Z. | | Deposit date: | 2023-05-05 | | Release date: | 2023-08-09 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Inhibitory mechanism of CRISPR-Cas9 by AcrIIC4.
Nucleic Acids Res., 51, 2023
|
|
8JIZ
 
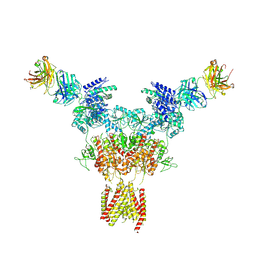 | |
8JJ2
 
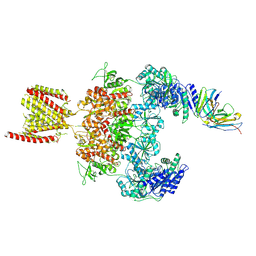 | | Cryo-EM structure of GluN1-2A NMDAR in complex with human Fab2G7 in one fab conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab2G7 Heavy Chain, ... | | Authors: | Wang, H, Zhu, S. | | Deposit date: | 2023-05-29 | | Release date: | 2024-06-05 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis for antibody-mediated NMDA receptor clustering and endocytosis in autoimmune encephalitis.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8JJ1
 
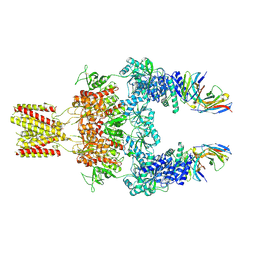 | | Cryo-EM structure of GluN1-2A NMDAR in complex with human Fab2G7 in two fab conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 2G7 Heavy Chain, ... | | Authors: | Wang, H, Zhu, S. | | Deposit date: | 2023-05-29 | | Release date: | 2024-06-05 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Structural basis for antibody-mediated NMDA receptor clustering and endocytosis in autoimmune encephalitis.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8JJ0
 
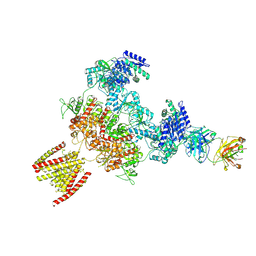 | |
8K4U
 
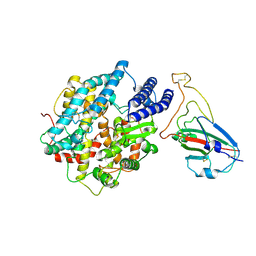 | |
5CDJ
 
 | | apical domain of chloroplast chaperonin 60a | | Descriptor: | RuBisCO large subunit-binding protein subunit alpha, chloroplastic | | Authors: | Zhang, S, Yu, F, Liu, C, Gao, F. | | Deposit date: | 2015-07-04 | | Release date: | 2016-07-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Functional Partition of Cpn60 alpha and Cpn60 beta Subunits in Substrate Recognition and Cooperation with Co-chaperonins
Mol Plant, 9, 2016
|
|
5CIO
 
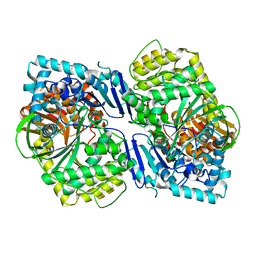 | | Crystal structure of PqqF | | Descriptor: | ZINC ION, pyrroloquinoline quinone biosynthesis protein PqqF | | Authors: | Wei, Q, Xu, D, Ran, T, Wang, W. | | Deposit date: | 2015-07-13 | | Release date: | 2016-06-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure and Function of PqqF Protein in the Pyrroloquinoline Quinone Biosynthetic Pathway
J.Biol.Chem., 291, 2016
|
|
5CDK
 
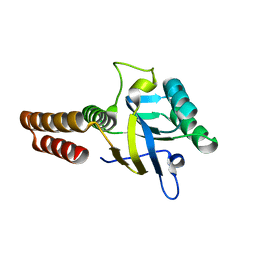 | | Apical domain of chloroplast chaperonin 60b1 | | Descriptor: | Chaperonin 60B1 | | Authors: | Zhang, S, Yu, F, Liu, C, Gao, F. | | Deposit date: | 2015-07-04 | | Release date: | 2016-07-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Functional Partition of Cpn60 alpha and Cpn60 beta Subunits in Substrate Recognition and Cooperation with Co-chaperonins
Mol Plant, 9, 2016
|
|
3UQR
 
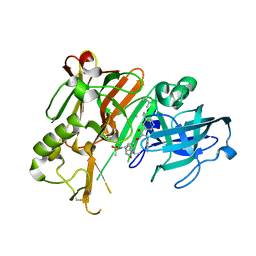 | | Crystal structure of BACE1 with its inhibitor | | Descriptor: | Beta-secretase 1, METHYL (2S)-1-[(2R,5S,8S,12S,13S)-2,13-DIBENZYL-12-HYDROXY-3,5-DIMETHYL-15-(3-[METHYL(METHYLSULFONYL)AMINO]-5-{[(1R)-1-PHENYLETHYL]CARBAMOYL}PHENYL)-8-(2-METHYLPROPYL)-4,7,10,15-TETRAOXO-3,6,9,14-TETRAAZAPENTADECAN-1-OYL]PYRROLIDINE-2-CARBOXYLATE | | Authors: | Chen, T.T, Chen, W.Y, Li, L, Xu, Y.C. | | Deposit date: | 2011-11-21 | | Release date: | 2012-11-21 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.056 Å) | | Cite: | Cyanobacterial Peptides as a Prototype for the Design of Potent beta-Secretase Inhibitors and the Development of Selective Chemical Probes for Other Aspartic Proteases
J.Med.Chem., 55, 2012
|
|
7W0D
 
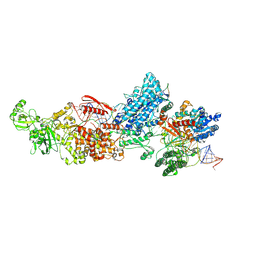 | | Dicer2-LoqsPD-dsRNA complex at mid-translocation state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Dicer-2, isoform A, ... | | Authors: | Su, S, Wang, J, Wang, H.W, Ma, J. | | Deposit date: | 2021-11-18 | | Release date: | 2022-04-27 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | Structural insights into dsRNA processing by Drosophila Dicer-2-Loqs-PD.
Nature, 607, 2022
|
|
7W0A
 
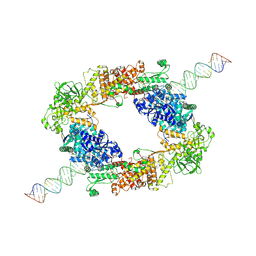 | | dmDicer2-LoqsPD-dsRNA Dimer status | | Descriptor: | Dicer-2, isoform A, Loquacious, ... | | Authors: | Su, S, Wang, J, Wang, H.W, Ma, J. | | Deposit date: | 2021-11-18 | | Release date: | 2022-04-27 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Structural insights into dsRNA processing by Drosophila Dicer-2-Loqs-PD.
Nature, 607, 2022
|
|
7W0C
 
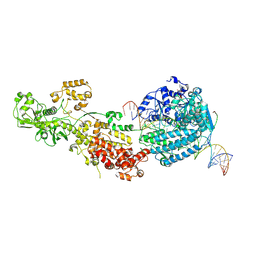 | | Dicer2-Loqs-PD-dsRNA complex at early-translocation state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Dicer-2, isoform A, ... | | Authors: | Su, S, Wang, J, Wang, H.W, Ma, J. | | Deposit date: | 2021-11-18 | | Release date: | 2022-04-27 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Structural insights into dsRNA processing by Drosophila Dicer-2-Loqs-PD.
Nature, 607, 2022
|
|
7W0E
 
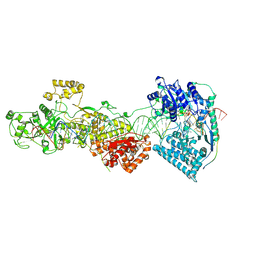 | | dmDicer2-LoqsPD-dsRNA Active-dicing status | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Dicer-2, isoform A, ... | | Authors: | Su, S, Wang, J, Wang, H.W, Ma, J. | | Deposit date: | 2021-11-18 | | Release date: | 2022-04-27 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (4.03 Å) | | Cite: | Structural insights into dsRNA processing by Drosophila Dicer-2-Loqs-PD.
Nature, 607, 2022
|
|
7W0B
 
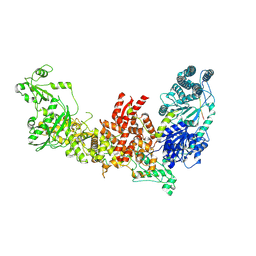 | | Dicer2-LoqsPD complex at apo status | | Descriptor: | Dicer-2, isoform A, Loquacious, ... | | Authors: | Su, S, Wang, J, Wang, H.W, Ma, J. | | Deposit date: | 2021-11-18 | | Release date: | 2022-04-27 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Structural insights into dsRNA processing by Drosophila Dicer-2-Loqs-PD.
Nature, 607, 2022
|
|
7W0F
 
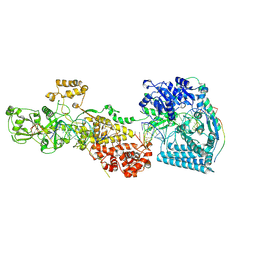 | | dmDicer2-LoqsPD-dsRNA Post-dicing status | | Descriptor: | Dicer-2, isoform A, Loquacious, ... | | Authors: | Su, S, Wang, J, Wang, H.W, Ma, J. | | Deposit date: | 2021-11-18 | | Release date: | 2022-04-27 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (4.55 Å) | | Cite: | Structural insights into dsRNA processing by Drosophila Dicer-2-Loqs-PD.
Nature, 607, 2022
|
|
8HI4
 
 | |
8HME
 
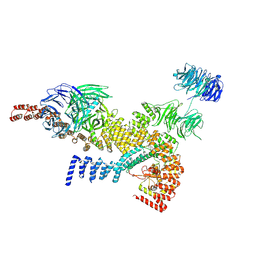 | | head module state 1 of Tetrahymena IFT-A | | Descriptor: | Intraflagellar transport protein 122 homolog, Intraflagellar transporter, WD40 repeat protein, ... | | Authors: | Ma, Y, Wu, J, Lei, M. | | Deposit date: | 2022-12-03 | | Release date: | 2023-06-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural insight into the intraflagellar transport complex IFT-A and its assembly in the anterograde IFT train.
Nat Commun, 14, 2023
|
|
8HMC
 
 | | base module state 1 of Tetrahymena IFT-A | | Descriptor: | Intraflagellar transport protein 122 homolog, Intraflagellar transport protein 43 homolog, Tetratricopeptide repeat protein, ... | | Authors: | Ma, Y, Wu, J, Lei, M. | | Deposit date: | 2022-12-03 | | Release date: | 2023-06-14 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insight into the intraflagellar transport complex IFT-A and its assembly in the anterograde IFT train.
Nat Commun, 14, 2023
|
|
6LNZ
 
 | |
