5XFA
 
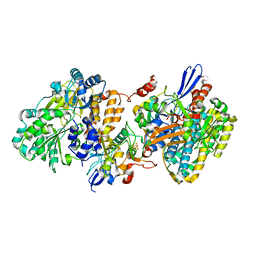 | | Crystal structure of NAD+-reducing [NiFe]-hydrogenase in the H2-reduced state | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE2/S2 (INORGANIC) CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Shomura, Y, Taketa, M, Nakashima, H, Tai, H, Nakagawa, H, Ikeda, Y, Ishii, M, Igarashi, Y, Nishihara, H, Yoon, K.S, Ogo, S, Hirota, S, Higuchi, Y. | | Deposit date: | 2017-04-09 | | Release date: | 2017-08-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of the redox switches in the NAD(+)-reducing soluble [NiFe]-hydrogenase
Science, 357, 2017
|
|
6ABU
 
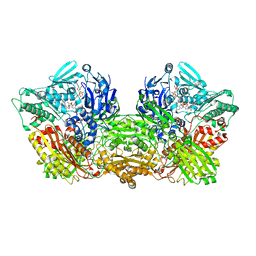 | | Rat Xanthine oxidoreductase, NAD bound form | | Descriptor: | BICARBONATE ION, FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Okamoto, K, Kawaguchi, Y. | | Deposit date: | 2018-07-24 | | Release date: | 2019-07-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Rat Xanthine oxidoreductase, NAD bound form
To Be Published
|
|
5XF9
 
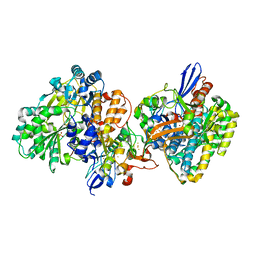 | | Crystal structure of NAD+-reducing [NiFe]-hydrogenase in the air-oxidized state | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Shomura, Y, Taketa, M, Nakashima, H, Tai, H, Nakagawa, H, Ikeda, Y, Ishii, M, Igarashi, Y, Nishihara, H, Yoon, K.S, Ogo, S, Hirota, S, Higuchi, Y. | | Deposit date: | 2017-04-09 | | Release date: | 2017-08-23 | | Last modified: | 2017-09-20 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural basis of the redox switches in the NAD(+)-reducing soluble [NiFe]-hydrogenase
Science, 357, 2017
|
|
7E5O
 
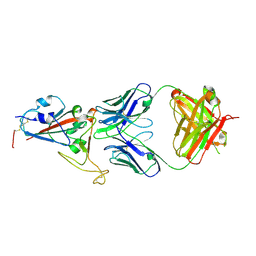 | | Crystal structure of SARS-CoV-2 RBD in complex with antibody NT-193 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NT-193 Heavy chain, NT-193 Light chain, ... | | Authors: | Kita, S, Onodera, T, Adachi, Y, Moriayma, S, Nomura, T, Tadokoro, T, Anraku, Y, Yumoto, K, Tian, C, Fukuhara, H, Suzuki, T, Tonouchi, K, Sasaki, J, Sun, L, Hashiguchi, T, Takahashi, Y, Maenaka, K. | | Deposit date: | 2021-02-19 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A SARS-CoV-2 antibody broadly neutralizes SARS-related coronaviruses and variants by coordinated recognition of a virus-vulnerable site.
Immunity, 54, 2021
|
|
1A7V
 
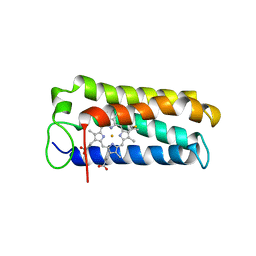 | | CYTOCHROME C' FROM RHODOPSEUDOMONAS PALUSTRIS | | Descriptor: | CYTOCHROME C', PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Shibata, N, Iba, S, Misaki, S, Meyer, T.E, Bartsch, R.G, Cusanovich, M.A, Higuchi, Y, Yasuoka, N. | | Deposit date: | 1998-03-18 | | Release date: | 1998-06-17 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Basis for monomer stabilization in Rhodopseudomonas palustris cytochrome c' derived from the crystal structure.
J.Mol.Biol., 284, 1998
|
|
3VZE
 
 | |
7YU1
 
 | |
7YU0
 
 | |
1J0P
 
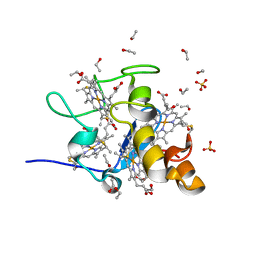 | | Three dimensional Structure of the Y43L mutant of Tetraheme Cytochrome c3 from Desulfovibrio vulgaris Miyazaki F | | Descriptor: | Cytochrome c3, ETHANOL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Ozawa, K, Yasukawa, F, Kumagai, J, Ohmura, T, Cusanvich, M.A, Tomimoto, Y, Ogata, H, Higuchi, Y, Akutsu, H. | | Deposit date: | 2002-11-19 | | Release date: | 2003-11-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (0.91 Å) | | Cite: | Role of the aromatic ring of Tyr43 in tetraheme cytochrome c(3) from Desulfovibrio vulgaris Miyazaki F.
Biophys.J., 85, 2003
|
|
5GZ9
 
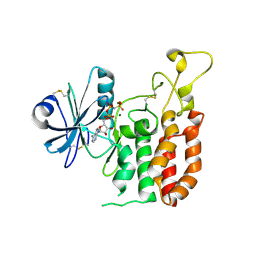 | | Crystal structure of catalytic domain of Protein O-mannosyl Kinase in complexes with AMP-PNP, Magnesium ions and glycopeptide | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Protein O-mannose kinase, ... | | Authors: | Nagae, M, Yamaguchi, Y. | | Deposit date: | 2016-09-27 | | Release date: | 2017-03-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 3D structural analysis of protein O-mannosyl kinase, POMK, a causative gene product of dystroglycanopathy.
Genes Cells, 22, 2017
|
|
7YU2
 
 | |
5GYR
 
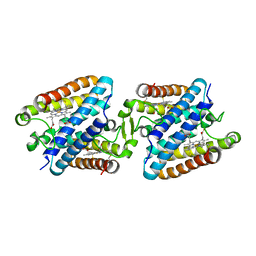 | | Tetrameric Allochromatium vinosum cytochrome c' | | Descriptor: | Cytochrome c', HEME C | | Authors: | Yamanaka, M, Hoshizumi, M, Nagao, S, Nakayama, R, Shibata, N, Higuchi, Y, Hirota, S. | | Deposit date: | 2016-09-23 | | Release date: | 2017-02-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Formation and carbon monoxide-dependent dissociation of Allochromatium vinosum cytochrome c' oligomers using domain-swapped dimers
Protein Sci., 26, 2017
|
|
3VY6
 
 | |
3VZF
 
 | |
3VZG
 
 | |
1J0O
 
 | | High Resolution Crystal Structure of the wild type Tetraheme Cytochrome c3 from Desulfovibrio vulgaris Miyazaki F | | Descriptor: | Cytochrome c3, ETHANOL, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ozawa, K, Yasukawa, F, Kumagai, J, Ohmura, T, Cusanvich, M.A, Tomimoto, Y, Ogata, H, Higuchi, Y, Akutsu, H. | | Deposit date: | 2002-11-19 | | Release date: | 2003-11-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Role of the aromatic ring of Tyr43 in tetraheme cytochrome c(3) from Desulfovibrio vulgaris Miyazaki F.
Biophys.J., 85, 2003
|
|
3WBQ
 
 | | Crystal structure of carbohydrate recognition domain of Blood Dendritic Cell Antigen-2 (BDCA2) lectin (crystal form-2) | | Descriptor: | C-type lectin domain family 4 member C | | Authors: | Nagae, M, Ikeda, A, Kitago, Y, Matsumoto, N, Yamamoto, K, Yamaguchi, Y. | | Deposit date: | 2013-05-20 | | Release date: | 2013-12-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of carbohydrate recognition domain of blood dendritic cell antigen-2 (BDCA2) reveal a common domain-swapped dimer.
Proteins, 82, 2014
|
|
3VY7
 
 | |
3WWK
 
 | | Crystal structure of CLEC-2 in complex with rhodocytin | | Descriptor: | C-type lectin domain family 1 member B, Snaclec rhodocytin subunit alpha, Snaclec rhodocytin subunit beta | | Authors: | Nagae, M, Morita-Matsumoto, K, Kato, M, Kato-Kaneko, M, Kato, Y, Yamaguchi, Y. | | Deposit date: | 2014-06-20 | | Release date: | 2014-10-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | A Platform of C-type Lectin-like Receptor CLEC-2 for Binding O-Glycosylated Podoplanin and Nonglycosylated Rhodocytin
Structure, 22, 2014
|
|
3WBR
 
 | | Crystal structure of carbohydrate recognition domain of Blood Dendritic Cell Antigen-2 (BDCA2) lectin (crystal form-3) | | Descriptor: | C-type lectin domain family 4 member C | | Authors: | Nagae, M, Ikeda, A, Kitago, Y, Matsumoto, N, Yamamoto, K, Yamaguchi, Y. | | Deposit date: | 2013-05-20 | | Release date: | 2013-12-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of carbohydrate recognition domain of blood dendritic cell antigen-2 (BDCA2) reveal a common domain-swapped dimer.
Proteins, 82, 2014
|
|
3WBP
 
 | | Crystal structure of carbohydrate recognition domain of Blood Dendritic Cell Antigen-2 (BDCA2) lectin (crystal form-1) | | Descriptor: | 1,2-ETHANEDIOL, C-type lectin domain family 4 member C | | Authors: | Nagae, M, Ikeda, A, Kitago, Y, Matsumoto, N, Yamamoto, K, Yamaguchi, Y. | | Deposit date: | 2013-05-20 | | Release date: | 2013-12-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of carbohydrate recognition domain of blood dendritic cell antigen-2 (BDCA2) reveal a common domain-swapped dimer.
Proteins, 82, 2014
|
|
3WSR
 
 | | Crystal structure of CLEC-2 in complex with O-glycosylated podoplanin | | Descriptor: | C-type lectin domain family 1 member B, Peptide from Podoplanin, beta-D-galactopyranose-(1-3)-[N-acetyl-alpha-neuraminic acid-(2-6)]2-acetamido-2-deoxy-alpha-D-galactopyranose | | Authors: | Nagae, M, Morita-Matsumoto, K, Kato, M, Kato-Kaneko, M, Kato, Y, Yamaguchi, Y. | | Deposit date: | 2014-03-20 | | Release date: | 2014-10-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | A Platform of C-type Lectin-like Receptor CLEC-2 for Binding O-Glycosylated Podoplanin and Nonglycosylated Rhodocytin
Structure, 22, 2014
|
|
7WCQ
 
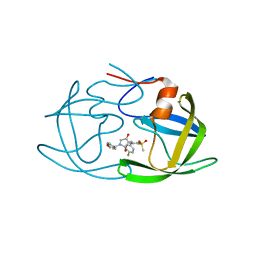 | | Crystal structure of HIV-1 protease in complex with lactam derivative 1 | | Descriptor: | (3R,4R)-3-[(4-fluorophenyl)methyl]-1-[(4-methoxyphenyl)methyl]-3-(4-methylsulfonylphenyl)-4-oxidanyl-pyrrolidin-2-one, Protease | | Authors: | Kojima, E, Iimuro, A, Nakajima, M, Kinuta, H, Asada, N, Sako, Y, Nakata, Z, Uemura, K, Arita, S, Miki, S, Wakasa-Morimoto, C, Tachibana, Y, Fumoto, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.011 Å) | | Cite: | Pocket-to-Lead: Structure-Based De Novo Design of Novel Non-peptidic HIV-1 Protease Inhibitors Using the Ligand Binding Pocket as a Template.
J.Med.Chem., 65, 2022
|
|
7WBS
 
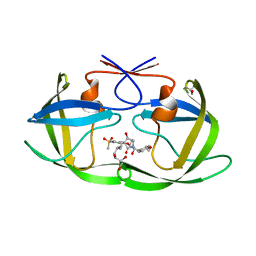 | | Crystal structure of HIV-1 protease in complex with lactam derivative 2 | | Descriptor: | (3~{R},4~{R})-1-[(4-methoxyphenyl)methyl]-3-(3-methylbutyl)-3-[4-methylsulfonyl-2-[(2~{S})-1-oxidanylpropan-2-yl]oxy-phenyl]-4-oxidanyl-pyrrolidin-2-one, GLYCEROL, Protease | | Authors: | Kojima, E, Iimuro, A, Nakajima, M, Kinuta, H, Asada, N, Sako, Y, Nakata, Z, Uemura, K, Arita, S, Miki, S, Wakabayashi-Morimoto, C, Tachibana, Y, Fumoto, M. | | Deposit date: | 2021-12-17 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Pocket-to-Lead: Structure-Based De Novo Design of Novel Non-peptidic HIV-1 Protease Inhibitors Using the Ligand Binding Pocket as a Template.
J.Med.Chem., 65, 2022
|
|
8J83
 
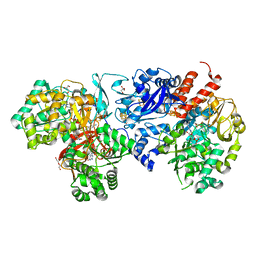 | | Crystal structure of formate dehydrogenase from Methylorubrum extorquens AM1 | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, DI(HYDROXYETHYL)ETHER, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Kobayashi, A, Taketa, M, Sowa, K, Kano, K, Higuchi, Y, Ogata, H. | | Deposit date: | 2023-04-29 | | Release date: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and function relationship of formate dehydrogenases: an overview of recent progress.
Iucrj, 10, 2023
|
|
