8JYA
 
 | | Crystal Structure of Intracellular B30.2 Domain of VpBTN3 in Complex with IPP | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, Butyrophylin 3, SULFATE ION | | Authors: | Yang, Y.Y, Yi, S.M, Zhang, M.T, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-07-03 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Phosphoantigens glue butyrophilin 3A1 and 2A1 to activate V gamma 9V delta 2 T cells.
Nature, 621, 2023
|
|
8JYF
 
 | | Crystal Structure of Intracellular B30.2 Domain of VpBTN3 in Complex with DMAPP | | Descriptor: | Butyrophylin 3, DIMETHYLALLYL DIPHOSPHATE, SULFATE ION | | Authors: | Yang, Y.Y, Yi, S.M, Zhang, M.T, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-07-03 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Phosphoantigens glue butyrophilin 3A1 and 2A1 to activate V gamma 9V delta 2 T cells.
Nature, 621, 2023
|
|
8JY9
 
 | | Crystal Structure of Intracellular B30.2 Domain of VpBTN3 in Complex with HMBPP | | Descriptor: | (2E)-4-hydroxy-3-methylbut-2-en-1-yl trihydrogen diphosphate, Butyrophylin 3, SULFATE ION | | Authors: | Yang, Y.Y, Yi, S.M, Zhang, M.T, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-07-03 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Phosphoantigens glue butyrophilin 3A1 and 2A1 to activate V gamma 9V delta 2 T cells.
Nature, 621, 2023
|
|
8JYB
 
 | | Crystal Structure of Intracellular B30.2 Domain of VpBTN3 | | Descriptor: | Butyrophylin 3, SULFATE ION | | Authors: | Yang, Y.Y, Yi, S.M, Zhang, M.T, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-07-03 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Phosphoantigens glue butyrophilin 3A1 and 2A1 to activate V gamma 9V delta 2 T cells.
Nature, 621, 2023
|
|
5GL5
 
 | |
4D3G
 
 | | Structure of PstA | | Descriptor: | PSTA | | Authors: | Campeotto, I, Freemont, P, Grundling, A. | | Deposit date: | 2014-10-22 | | Release date: | 2014-12-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Complex Structure and Biochemical Characterization of the Staphylococcus Aureus Cyclic Di-AMP Binding Protein Psta, the Founding Member of a New Signal Transduction Protein Family
J.Biol.Chem., 290, 2015
|
|
5XVJ
 
 | | Crystal structure of AL7 PAL domain | | Descriptor: | PHD finger protein ALFIN-LIKE 7, SULFATE ION | | Authors: | Peng, L, Wang, L.L, Huang, Y. | | Deposit date: | 2017-06-28 | | Release date: | 2018-07-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Analysis of the Arabidopsis AL2-PAL and PRC1 Complex Provides Mechanistic Insight into Active-to-Repressive Chromatin State Switch
J. Mol. Biol., 430, 2018
|
|
5XVM
 
 | |
4DIE
 
 | |
4BHK
 
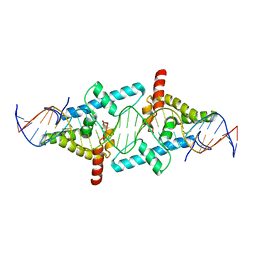 | | Crystal Structure of Moss Leafy bound to DNA | | Descriptor: | FLORICAULA/LEAFY HOMOLOG 1, MOSS-CR54 DNA | | Authors: | Nanao, M.H, Sayou, C, Dumas, R, Parcy, F. | | Deposit date: | 2013-04-03 | | Release date: | 2014-01-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | A Promiscuous Intermediate Underlies the Evolution of Leafy DNA Binding Specificity.
Science, 343, 2014
|
|
5XVL
 
 | | Crystal structure of AL2 PAL domain | | Descriptor: | PHD finger protein ALFIN-LIKE 2, SULFATE ION | | Authors: | Peng, L, Wang, L.L, Huang, Y. | | Deposit date: | 2017-06-28 | | Release date: | 2018-07-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.629 Å) | | Cite: | Structural Analysis of the Arabidopsis AL2-PAL and PRC1 Complex Provides Mechanistic Insight into Active-to-Repressive Chromatin State Switch
J. Mol. Biol., 430, 2018
|
|
4EGF
 
 | |
1CHZ
 
 | | A NEW NEUROTOXIN FROM BUTHUS MARTENSII KARSCH | | Descriptor: | CHLORIDE ION, PROTEIN (BMK M2) | | Authors: | He, X.L, Deng, J.P, Li, H.M, Wang, D.C. | | Deposit date: | 1999-03-31 | | Release date: | 2000-03-31 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structure of a new neurotoxin from the scorpion Buthus martensii Karsch at 1.76 A.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
4ERS
 
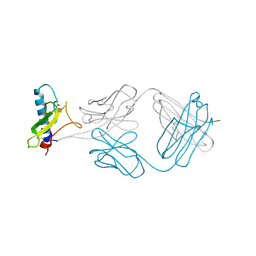 | | A Molecular Basis for Negative Regulation of the Glucagon Receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab heavy chain, Fab light chain, ... | | Authors: | Murray, J.M, Koth, C.M, Mukund, S. | | Deposit date: | 2012-04-20 | | Release date: | 2012-08-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.637 Å) | | Cite: | Molecular basis for negative regulation of the glucagon receptor.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4DI1
 
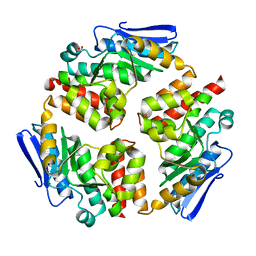 | |
4EYE
 
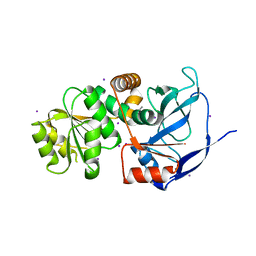 | |
4E98
 
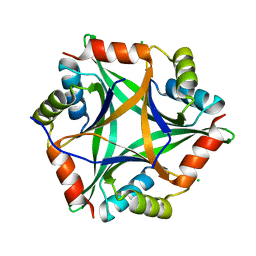 | |
4ED4
 
 | |
4EO9
 
 | |
4EGE
 
 | |
4EMD
 
 | |
6LUM
 
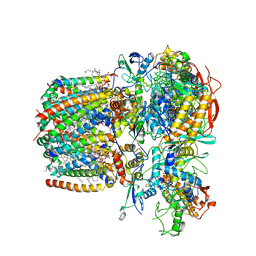 | | Structure of Mycobacterium smegmatis succinate dehydrogenase 2 | | Descriptor: | (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOINOSITOL, 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, ... | | Authors: | Gao, Y, Gong, H, Zhou, X, Xiao, Y, Wang, W, Ji, W, Wang, Q, Rao, Z. | | Deposit date: | 2020-01-29 | | Release date: | 2020-05-27 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Cryo-EM structure of trimeric Mycobacterium smegmatis succinate dehydrogenase with a membrane-anchor SdhF.
Nat Commun, 11, 2020
|
|
7E3L
 
 | | Ultrapotent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 58G6 heavy chain, 58G6 light chain, ... | | Authors: | Guo, H, Li, T, Liu, F, Gao, Y, Ji, X, Yang, H. | | Deposit date: | 2021-02-09 | | Release date: | 2021-09-15 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Potent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants.
Nat Commun, 12, 2021
|
|
7E3K
 
 | | Ultrapotent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants | | Descriptor: | 13G9 heavy chain, 13G9 light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, H, Li, T, Liu, F, Gao, Y, Ji, X, Yang, H. | | Deposit date: | 2021-02-09 | | Release date: | 2021-09-15 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Potent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants.
Nat Commun, 12, 2021
|
|
5Y6G
 
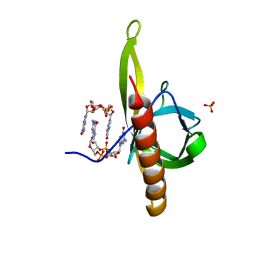 | | PilZ domain with c-di-GMP of YcgR from Escherichia coli | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Flagellar brake protein YcgR, SULFATE ION | | Authors: | Hou, Y.J, Wang, D.C, Li, D.F. | | Deposit date: | 2017-08-11 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the mechanism of c-di-GMP-bound YcgR regulating flagellar motility inEscherichia coli.
J.Biol.Chem., 295, 2020
|
|
