7CD9
 
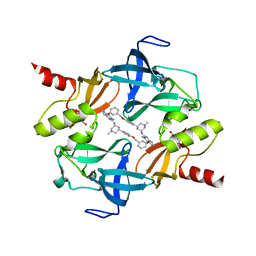 | | Crystal Structure of SETDB1 tudor domain in complexed with Compound 6 | | Descriptor: | 3-methyl-2-[[(3R,5R)-1-methyl-5-(4-phenylmethoxyphenyl)piperidin-3-yl]amino]-5H-pyrrolo[3,2-d]pyrimidin-4-one, CITRIC ACID, Histone-lysine N-methyltransferase SETDB1 | | Authors: | Xiong, L, Guo, Y, Mao, X, Huang, L, Wu, C, Yang, S. | | Deposit date: | 2020-06-19 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Guided Discovery of a Potent and Selective Cell-Active Inhibitor of SETDB1 Tudor Domain.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7CAJ
 
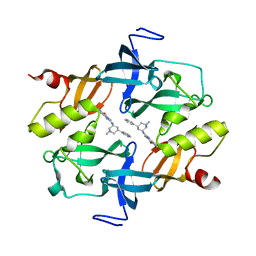 | | Crystal structure of SETDB1 Tudor domain in complexed with Compound 2. | | Descriptor: | 3-methyl-2-[[(3R,5R)-1-methyl-5-phenyl-piperidin-3-yl]amino]-5H-pyrrolo[3,2-d]pyrimidin-4-one, Histone-lysine N-methyltransferase SETDB1 | | Authors: | Guo, Y.P, Liang, X, Xin, M, Luyi, H, Chengyong, W, Yang, S.Y. | | Deposit date: | 2020-06-08 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Structure-Guided Discovery of a Potent and Selective Cell-Active Inhibitor of SETDB1 Tudor Domain.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
1ALW
 
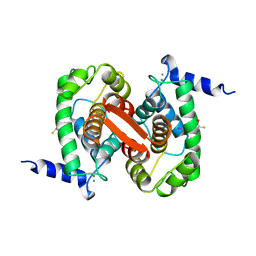 | | INHIBITOR AND CALCIUM BOUND DOMAIN VI OF PORCINE CALPAIN | | Descriptor: | 3-(4-IODO-PHENYL)-2-MERCAPTO-PROPIONIC ACID, CALCIUM ION, CALPAIN | | Authors: | Narayana, S.V.L, Lin, G. | | Deposit date: | 1997-06-04 | | Release date: | 1998-06-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structure of calcium bound domain VI of calpain at 1.9 A resolution and its role in enzyme assembly, regulation, and inhibitor binding.
Nat.Struct.Biol., 4, 1997
|
|
1ALV
 
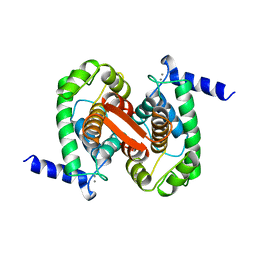 | | CALCIUM BOUND DOMAIN VI OF PORCINE CALPAIN | | Descriptor: | CALCIUM ION, CALPAIN | | Authors: | Narayana, S.V.L, Lin, G, Chattopadhyay, D, Maki, M. | | Deposit date: | 1997-06-03 | | Release date: | 1998-06-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of calcium bound domain VI of calpain at 1.9 A resolution and its role in enzyme assembly, regulation, and inhibitor binding.
Nat.Struct.Biol., 4, 1997
|
|
7F8H
 
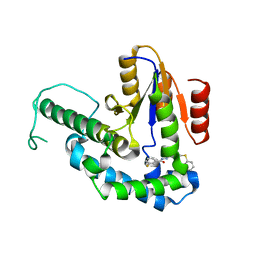 | |
7F8G
 
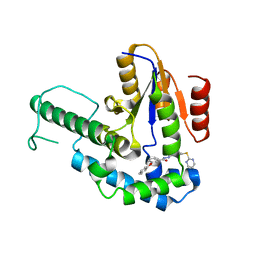 | |
2H42
 
 | | Crystal structure of PDE5 in complex with sildenafil | | Descriptor: | 5-{2-ETHOXY-5-[(4-METHYLPIPERAZIN-1-YL)SULFONYL]PHENYL}-1-METHYL-3-PROPYL-1H,6H,7H-PYRAZOLO[4,3-D]PYRIMIDIN-7-ONE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Wang, H, Ke, H. | | Deposit date: | 2006-05-23 | | Release date: | 2006-06-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Multiple Conformations of Phosphodiesterase-5: Implications for enzyme function and drug development
J.Biol.Chem., 281, 2006
|
|
2H44
 
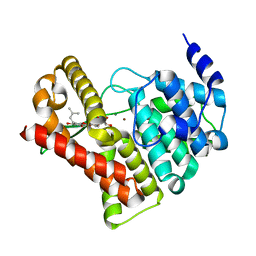 | | Crystal structure of PDE5A1 in complex with icarisid II | | Descriptor: | 5,7-DIHYDROXY-2-(4-METHOXYPHENYL)-8-(3-METHYLBUTYL)-4-OXO-4H-CHROMEN-3-YL 6-DEOXY-ALPHA-L-MANNOPYRANOSIDE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Wang, H, Ke, H. | | Deposit date: | 2006-05-23 | | Release date: | 2006-06-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Multiple Conformations of Phosphodiesterase-5: Implications for enzyme function and drug development
J.Biol.Chem., 281, 2006
|
|
8ETR
 
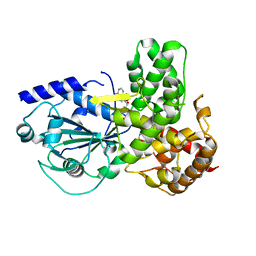 | | CryoEM Structure of NLRP3 NACHT domain in complex with G2394 | | Descriptor: | (6S,8R)-N-[(1,2,3,5,6,7-hexahydro-s-indacen-4-yl)carbamoyl]-6-(methylamino)-6,7-dihydro-5H-pyrazolo[5,1-b][1,3]oxazine-3-sulfonamide, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Murray, J.M, Johnson, M.C. | | Deposit date: | 2022-10-17 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Overcoming Preclinical Safety Obstacles to Discover ( S )- N -((1,2,3,5,6,7-Hexahydro- s -indacen-4-yl)carbamoyl)-6-(methylamino)-6,7-dihydro-5 H -pyrazolo[5,1- b ][1,3]oxazine-3-sulfonamide (GDC-2394): A Potent and Selective NLRP3 Inhibitor.
J.Med.Chem., 65, 2022
|
|
7B3E
 
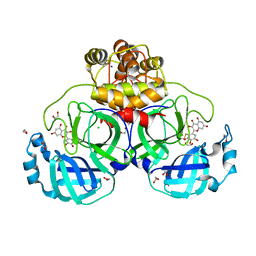 | | Crystal structure of myricetin covalently bound to the main protease (3CLpro/Mpro) of SARS-CoV-2 | | Descriptor: | 1,2-ETHANEDIOL, 3,5,7-TRIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, CHLORIDE ION, ... | | Authors: | Costanzi, E, Demitri, N, Giabbai, B, Storici, P. | | Deposit date: | 2020-11-30 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Identification of Inhibitors of SARS-CoV-2 3CL-Pro Enzymatic Activity Using a Small Molecule in Vitro Repurposing Screen.
Acs Pharmacol Transl Sci, 4, 2021
|
|
6GRZ
 
 | |
8F2Q
 
 | |
4UZD
 
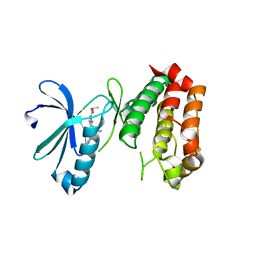 | | SAR156497 an exquisitely selective inhibitor of Aurora kinases | | Descriptor: | AURORA KINASE A, ethyl (9S)-9-[3-(1H-benzimidazol-2-yloxy)phenyl]-8-oxo-4,5,6,7,8,9-hexahydro-2H-pyrrolo[3,4-b]quinoline-3-carboxylate | | Authors: | Pouzieux, S, Delarbre, L, Crenne, J.Y. | | Deposit date: | 2014-09-05 | | Release date: | 2014-11-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Sar156497 an Exquisitely Selective Inhibitor of Aurora Kinases.
J.Med.Chem., 58, 2015
|
|
4UZH
 
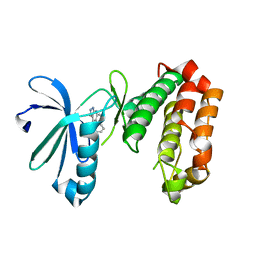 | | SAR156497 an exquisitely selective inhibitor of Aurora kinases | | Descriptor: | (4S)-4-(2-fluorophenyl)-2,4,6,7,8,9-hexahydro-5H-pyrazolo[3,4-b][1,7]naphthyridin-5-one, AURORA 2 KINASE DOMAIN | | Authors: | Pouzieux, S, Maignan, S, Crenne, J.Y. | | Deposit date: | 2014-09-05 | | Release date: | 2014-11-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Sar156497 an Exquisitely Selective Inhibitor of Aurora Kinases.
J.Med.Chem., 58, 2015
|
|
5LL9
 
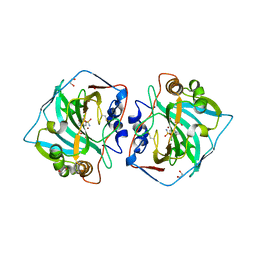 | | Crystal structure of human carbonic anhydrase isozyme XII with 4-(1H-benzimidazol-1-ylacetyl)-2-chlorobenzenesulfonamide | | Descriptor: | 1,2-ETHANEDIOL, 4-[2-(benzimidazol-1-yl)ethanoyl]-2-chloranyl-benzenesulfonamide, Carbonic anhydrase 12, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2016-07-27 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Combinatorial Design of Isoform-Selective N-Alkylated Benzimidazole-Based Inhibitors of Carbonic Anhydrases
Chemistryselect, 2017
|
|
5LLA
 
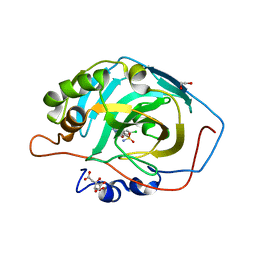 | | Crystal structure of human carbonic anhydrase isozyme XIII with 4-(1H-benzimidazol-1-ylacetyl)-2-chlorobenzenesulfonamide | | Descriptor: | 1,2-ETHANEDIOL, 4-[2-(benzimidazol-1-yl)ethanoyl]-2-chloranyl-benzenesulfonamide, CITRIC ACID, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2016-07-27 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Combinatorial Design of Isoform-Selective N-Alkylated Benzimidazole-Based Inhibitors of Carbonic Anhydrases
Chemistryselect, 2017
|
|
5LL5
 
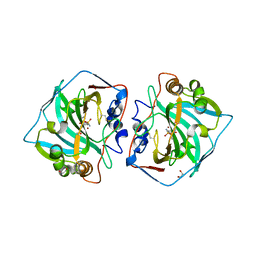 | | Crystal structure of human carbonic anhydrase isozyme XII with 4-(1H-benzimidazol-1-ylacetyl)benzenesulfonamide | | Descriptor: | 1,2-ETHANEDIOL, 4-[2-(benzimidazol-1-yl)ethanoyl]benzenesulfonamide, Carbonic anhydrase 12, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2016-07-26 | | Release date: | 2017-08-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Combinatorial Design of Isoform-Selective N-Alkylated Benzimidazole-Based Inhibitors of Carbonic Anhydrases
Chemistryselect, 2017
|
|
5LL4
 
 | |
3MON
 
 | |
6YRP
 
 | | Crystal Structure of the VIM-2 Acquired Metallo-beta-Lactamase in Complex with JMV-4690 (Cpd 31) | | Descriptor: | 1,2-ETHANEDIOL, 2-[[[3-(5-methoxy-2-oxidanyl-phenyl)-5-sulfanylidene-1~{H}-1,2,4-triazol-4-yl]amino]methyl]benzoic acid, ACETATE ION, ... | | Authors: | Docquier, J.D, Pozzi, C, De Luca, F, Benvenuti, M, Mangani, S. | | Deposit date: | 2020-04-20 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 4-Amino-1,2,4-triazole-3-thione-derived Schiff bases as metallo-beta-lactamase inhibitors.
Eur.J.Med.Chem., 208, 2020
|
|
6H96
 
 | | AlbA-albicidin complex, albicidin resistance protein | | Descriptor: | 4-[[4-[[4-[(3~{S})-5-azanyl-3-[[4-[[(~{E})-3-(4-hydroxyphenyl)-2-methyl-prop-2-enoyl]amino]phenyl]carbonylamino]-2-oxidanylidene-3~{H}-pyrrol-1-yl]phenyl]carbonylamino]-3-methoxy-2-oxidanyl-phenyl]carbonylamino]-3-methoxy-2-oxidanyl-benzoic acid, Albicidin resistance protein, SULFATE ION | | Authors: | Koehnke, J, Sikandar, A. | | Deposit date: | 2018-08-03 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Adaptation of a Bacterial Multidrug Resistance System Revealed by the Structure and Function of AlbA.
J.Am.Chem.Soc., 140, 2018
|
|
6H95
 
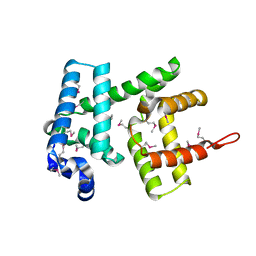 | | AlbA, albicidin resistance protein | | Descriptor: | Albicidin resistance protein | | Authors: | Koehnke, J, Sikandar, A. | | Deposit date: | 2018-08-03 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Adaptation of a Bacterial Multidrug Resistance System Revealed by the Structure and Function of AlbA.
J.Am.Chem.Soc., 140, 2018
|
|
6V3P
 
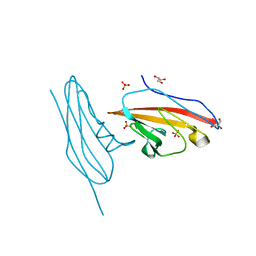 | |
1PGV
 
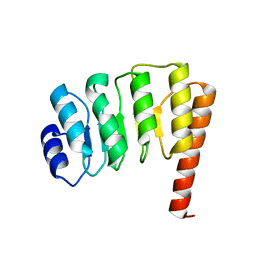 | | Structural Genomics of Caenorhabditis elegans: tropomodulin C-terminal domain | | Descriptor: | tropomodulin TMD-1 | | Authors: | Symersky, J, Lu, S, Li, S, Chen, L, Meehan, E, Luo, M, Qiu, S, Bunzel, R.J, Luo, D, Arabashi, A, Nagy, L.A, Lin, G, Luan, W.C.-H, Carson, M, Gray, R, Huang, W, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-05-28 | | Release date: | 2003-06-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural genomics of Caenorhabditis elegans: crystal structure of the tropomodulin C-terminal domain
Proteins, 56, 2004
|
|
1OOE
 
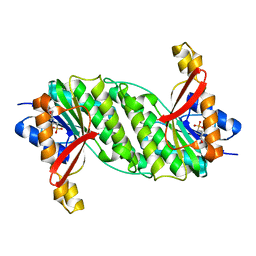 | | Structural Genomics of Caenorhabditis elegans : Dihydropteridine reductase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Dihydropteridine reductase | | Authors: | Symersky, J, Li, S, Nagy, L, Qiu, S, Lin, G, Tsao, J, Luo, D, Carson, M, DeLucas, L, Luo, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-03-03 | | Release date: | 2003-03-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural genomics of Caenorhabditis elegans: structure of dihydropteridine reductase.
Proteins, 53, 2003
|
|
