1PF5
 
 | | Structural Genomics, Protein YJGH | | Descriptor: | Hypothetical protein yjgH, MERCURY (II) ION | | Authors: | Zhang, R, Joachimiak, A, Edwards, A, Savchenko, A, Xu, L, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-05-23 | | Release date: | 2003-12-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The 2.5A crystal structure of protein YJGH from E. Coli
To be Published
|
|
1TP6
 
 | | 1.5 A Crystal Structure of a NTF-2 Like Protein of Unknown Function PA1314 from Pseudomonas aeruginosa | | Descriptor: | hypothetical protein PA1314 | | Authors: | Zhang, R, Xu, L.X, savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-06-15 | | Release date: | 2004-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.5A crystal structure of a hypothetical protein PA1314 from Pseudomonas aeruginosa
To be Published
|
|
1TUA
 
 | | 1.5 A Crystal Structure of a Protein of Unknown Function APE0754 from Aeropyrum pernix | | Descriptor: | Hypothetical protein APE0754 | | Authors: | Zhang, R, Skarina, T, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-06-24 | | Release date: | 2004-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.5A crystal structure of a hypothetical protein APE0754 from Aeropyrum pernix
To be Published
|
|
2HRZ
 
 | | The crystal structure of the nucleoside-diphosphate-sugar epimerase from Agrobacterium tumefaciens | | Descriptor: | Nucleoside-diphosphate-sugar epimerase | | Authors: | Zhang, R, Xu, X, Zheng, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-07-20 | | Release date: | 2006-08-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The crystal structure of the nucleoside-diphosphate-sugar epimerase from Agrobacterium tumefaciens
To be Published
|
|
2I0M
 
 | | Crystal structure of the phosphate transport system regulatory protein PhoU from Streptococcus pneumoniae | | Descriptor: | Phosphate transport system protein phoU, ZINC ION | | Authors: | Zhang, R, Li, H, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-08-10 | | Release date: | 2006-09-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the phosphate transport system regulatory protein PhoU from Streptococcus pneumoniae
To be Published, 2006
|
|
2IAZ
 
 | |
7VEP
 
 | |
1EUI
 
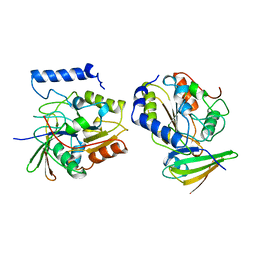 | | ESCHERICHIA COLI URACIL-DNA GLYCOSYLASE COMPLEX WITH URACIL-DNA GLYCOSYLASE INHIBITOR PROTEIN | | Descriptor: | URACIL-DNA GLYCOSYLASE, URACIL-DNA GLYCOSYLASE INHIBITOR PROTEIN | | Authors: | Ravishankar, R, Sagar, M.B, Roy, S, Purnapatre, K, Handa, P, Varshney, U, Vijayan, M. | | Deposit date: | 1998-06-18 | | Release date: | 1999-06-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | X-ray analysis of a complex of Escherichia coli uracil DNA glycosylase (EcUDG) with a proteinaceous inhibitor. The structure elucidation of a prokaryotic UDG.
Nucleic Acids Res., 26, 1998
|
|
6B0V
 
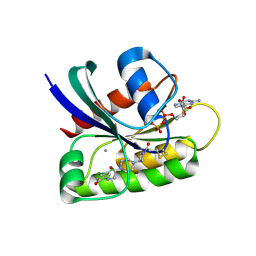 | | Crystal Structure of small molecule ARS-107 covalently bound to K-Ras G12C | | Descriptor: | 1-[3-(4-{[(4,5-dichloro-2-hydroxyphenyl)amino]acetyl}piperazin-1-yl)azetidin-1-yl]propan-1-one, CALCIUM ION, GTPase KRas, ... | | Authors: | Hansen, R, Peters, U, Babbar, A, Chen, Y, Feng, J, Janes, M.R, Li, L.-S, Ren, P, Liu, Y, Zarrinkar, P.P. | | Deposit date: | 2017-09-15 | | Release date: | 2018-05-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | The reactivity-driven biochemical mechanism of covalent KRASG12Cinhibitors.
Nat. Struct. Mol. Biol., 25, 2018
|
|
3U7I
 
 | | The crystal structure of FMN-dependent NADH-azoreductase 1 (GBAA0966) from Bacillus anthracis str. Ames Ancestor | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, FMN-dependent NADH-azoreductase 1, ... | | Authors: | Zhang, R, Gu, M, Tan, K, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-10-13 | | Release date: | 2011-11-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The crystal structure of FMN-dependent NADH-azoreductase 1 (GBAA0966) from Bacillus anthracis str. Ames Ancestor
To be Published
|
|
6B0Y
 
 | | Crystal Structure of small molecule ARS-917 covalently bound to K-Ras G12C | | Descriptor: | 1-{4-[6-chloro-7-(2-fluorophenyl)quinazolin-4-yl]piperazin-1-yl}propan-1-one, CALCIUM ION, GLYCEROL, ... | | Authors: | Hansen, R, Peters, U, Babbar, A, Chen, Y, Feng, J, Janes, M.R, Li, L.-S, Ren, P, Liu, Y, Zarrinkar, P.P. | | Deposit date: | 2017-09-15 | | Release date: | 2018-05-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | The reactivity-driven biochemical mechanism of covalent KRASG12Cinhibitors.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6BJC
 
 | | TPX2_mini decorated GMPCPP-microtubule | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, ... | | Authors: | Zhang, R, Nogales, E. | | Deposit date: | 2017-11-05 | | Release date: | 2017-11-22 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insight into TPX2-stimulated microtubule assembly.
Elife, 6, 2017
|
|
3OSX
 
 | |
1PLY
 
 | | SODIUM IONS AND WATER MOLECULES IN THE STRUCTURE OF POLY D(A)(DOT)POLY D(T) | | Descriptor: | DNA (5'-D(P*AP*AP*AP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*TP*T)-3'), SODIUM ION | | Authors: | Chandrasekaran, R, Radha, A, Park, H.-S. | | Deposit date: | 1995-02-28 | | Release date: | 1995-06-03 | | Last modified: | 2024-02-14 | | Method: | FIBER DIFFRACTION (3.2 Å) | | Cite: | Sodium ions and water molecules in the structure of poly(dA).poly(dT).
Acta Crystallogr.,Sect.D, 51, 1995
|
|
6IFE
 
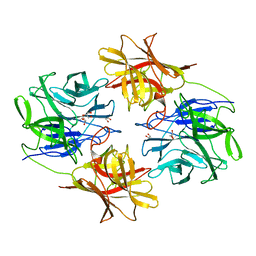 | | A Glycoside Hydrolase Family 43 beta-Xylosidase | | Descriptor: | Beta-xylosidase, GLYCEROL | | Authors: | Li, N, Liu, Y, Zhang, R, Zhou, J.P, Huang, Z.X. | | Deposit date: | 2018-09-20 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.804 Å) | | Cite: | Biochemical and structural properties of a low-temperature-active glycoside hydrolase family 43 beta-xylosidase: Activity and instability at high neutral salt concentrations.
Food Chem, 301, 2019
|
|
6OMX
 
 | |
6UV7
 
 | |
6UVI
 
 | |
5W1E
 
 | | PobR in complex with PHB | | Descriptor: | GLYCEROL, P-HYDROXYBENZOIC ACID, Putative transcriptional regulator, ... | | Authors: | Page, R, Peti, W, Lord, D.M, Bajaj, R, Zhang, R. | | Deposit date: | 2017-06-02 | | Release date: | 2017-12-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | A peculiar IclR family transcription factor regulates para-hydroxybenzoate catabolism in Streptomyces coelicolor.
Nucleic Acids Res., 46, 2018
|
|
6LW8
 
 | | Structural basis for domain rotation during adenylation of active site K123 and fragment library screening against NAD+ -dependent DNA ligase from Mycobacterium tuberculosis | | Descriptor: | (4R)-4-(4-fluorophenyl)-4,5,6,7-tetrahydro-1H-imidazo[4,5-c]pyridine, DNA ligase A, GLYCEROL, ... | | Authors: | Ramachandran, R, Afsar, M, Shukla, A. | | Deposit date: | 2020-02-07 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structure based identification of first-in-class fragment inhibitors that target the NMN pocket of M. tuberculosis NAD + -dependent DNA ligase A.
J.Struct.Biol., 213, 2021
|
|
8K4E
 
 | | Cryo-EM structure of 30S ribosome with cleaved AP-mRNA bound complex-II | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Ramachandran, R, Afsar, M, Shukla, A. | | Deposit date: | 2023-07-18 | | Release date: | 2024-07-24 | | Last modified: | 2025-01-08 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Bacterial Rps3 counters oxidative and UV stress by recognizing and processing AP-sites on mRNA via a novel mechanism.
Nucleic Acids Res., 52, 2024
|
|
3FFA
 
 | | Crystal Structure of a fast activating G protein mutant | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Guanine nucleotide-binding protein G(i), alpha-1 subunit, ... | | Authors: | Chauhan, R, Kapoor, N. | | Deposit date: | 2008-12-02 | | Release date: | 2009-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural evidence for a sequential release mechanism for activation of heterotrimeric g proteins.
J.Mol.Biol., 393, 2009
|
|
3FFB
 
 | | Gi-alpha-1 mutant in GDP bound form | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(i), alpha-1 subunit, ... | | Authors: | Chauhan, R, Kapoor, N. | | Deposit date: | 2008-12-02 | | Release date: | 2009-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structural evidence for a sequential release mechanism for activation of heterotrimeric g proteins.
J.Mol.Biol., 393, 2009
|
|
3PYW
 
 | | The structure of the SLH domain from B. anthracis surface array protein at 1.8A | | Descriptor: | S-layer protein sap, SULFATE ION | | Authors: | Zhang, R, Wilton, R, Kern, J, Joachimiak, A, Schneewind, O, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-12-13 | | Release date: | 2011-04-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Surface Layer Homology (SLH) Domains from Bacillus anthracis Surface Array Protein.
J.Biol.Chem., 286, 2011
|
|
1XBW
 
 | | 1.9A Crystal Structure of the protein isdG from Staphylococcus aureus aureus, Structural genomics, MCSG | | Descriptor: | hypothetical protein isdG | | Authors: | Zhang, R, Wu, R, Joachimiak, G, Schneewind, O, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-08-31 | | Release date: | 2004-10-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Staphylococcus aureus IsdG and IsdI, heme-degrading enzymes with structural similarity to monooxygenases.
J.Biol.Chem., 280, 2005
|
|
