8H5C
 
 | | Structure of SARS-CoV-2 Omicron BA.2.75 RBD in complex with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Z.N, Bai, B, Liu, K.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2022-10-12 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for receptor binding and broader interspecies receptor recognition of currently circulating Omicron sub-variants.
Nat Commun, 14, 2023
|
|
6JO7
 
 | | Crystal structure of mouse MXRA8 | | Descriptor: | Matrix remodeling-associated protein 8 | | Authors: | Song, H, Zhao, Z, Qi, J, Gao, F, Gao, G.F. | | Deposit date: | 2019-03-20 | | Release date: | 2019-05-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular Basis of Arthritogenic Alphavirus Receptor MXRA8 Binding to Chikungunya Virus Envelope Protein.
Cell, 177, 2019
|
|
6JO8
 
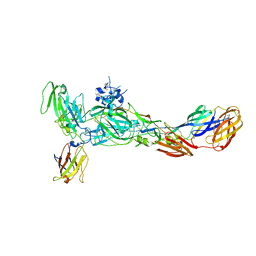 | | The complex structure of CHIKV envelope glycoprotein bound to human MXRA8 | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHIKV E1, ... | | Authors: | Song, H, Zhao, Z, Qi, J, Gao, F, Gao, F.G. | | Deposit date: | 2019-03-20 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.495 Å) | | Cite: | Molecular Basis of Arthritogenic Alphavirus Receptor MXRA8 Binding to Chikungunya Virus Envelope Protein.
Cell, 177, 2019
|
|
8HPU
 
 | |
8HQ7
 
 | |
8HPF
 
 | |
4Y9L
 
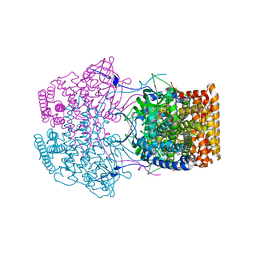 | | Crystal Structure of Caenorhabditis elegans ACDH-11 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Protein ACDH-11, isoform b | | Authors: | Li, Z.J, Zhai, Y.J, Zhang, K, Sun, F. | | Deposit date: | 2015-02-17 | | Release date: | 2015-06-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Acyl-CoA Dehydrogenase Drives Heat Adaptation by Sequestering Fatty Acids
Cell, 161, 2015
|
|
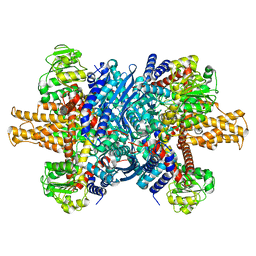
7VDA
 
 | |
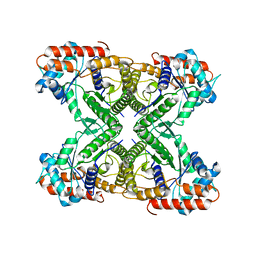
7VDC
 
 | |
7VD8
 
 | |
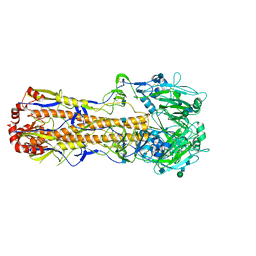
7VDF
 
 | |
7VD9
 
 | | 2.29 A structure of the human catalase | | Descriptor: | Catalase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Fan, H.C, Zhang, Y, Sun, F. | | Deposit date: | 2021-09-06 | | Release date: | 2021-12-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.29 Å) | | Cite: | A cryo-electron microscopy support film formed by 2D crystals of hydrophobin HFBI.
Nat Commun, 12, 2021
|
|
7VDE
 
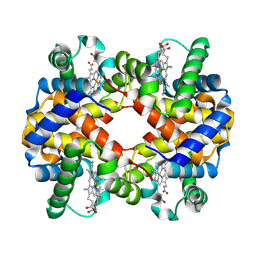 | | 3.6 A structure of the human hemoglobin | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Fan, H.C, Zhang, Y, Sun, F. | | Deposit date: | 2021-09-06 | | Release date: | 2021-12-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A cryo-electron microscopy support film formed by 2D crystals of hydrophobin HFBI.
Nat Commun, 12, 2021
|
|
6IEC
 
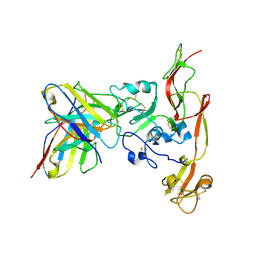 | | Structure of RVFV Gn and human monoclonal antibody R17 | | Descriptor: | NSmGnGc, R17 H chain, R17 L chain | | Authors: | Wang, Q.H, Wu, Y, Gao, F, Qi, J.X, Gao, G.F. | | Deposit date: | 2018-09-13 | | Release date: | 2019-04-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Neutralization mechanism of human monoclonal antibodies against Rift Valley fever virus.
Nat Microbiol, 4, 2019
|
|
6IEA
 
 | | Structure of RVFV Gn and human monoclonal antibody R13 | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, NSmGnGc, ... | | Authors: | Wang, Q.H, Wu, Y, Gao, F, Qi, J.X, Gao, G.F. | | Deposit date: | 2018-09-13 | | Release date: | 2019-04-10 | | Last modified: | 2019-07-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Neutralization mechanism of human monoclonal antibodies against Rift Valley fever virus.
Nat Microbiol, 4, 2019
|
|
6IEK
 
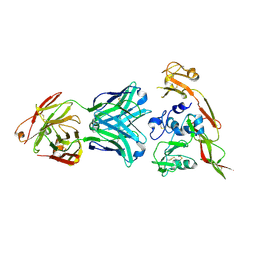 | | Structure of RVFV Gn and human monoclonal antibody R12 | | Descriptor: | Heavy chain of Fab R12, Light chain of Fab R12, NSmGnGc | | Authors: | Wang, Q.H, Wu, Y, Gao, F, Qi, J.X, Gao, G.F. | | Deposit date: | 2018-09-14 | | Release date: | 2019-04-10 | | Last modified: | 2019-07-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Neutralization mechanism of human monoclonal antibodies against Rift Valley fever virus.
Nat Microbiol, 4, 2019
|
|
6IEB
 
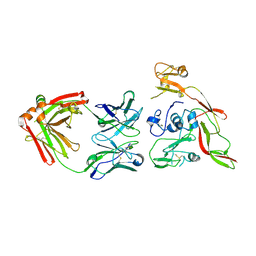 | | Structure of RVFV Gn and human monoclonal antibody R15 | | Descriptor: | NSmGnGc, R15 H chain, R15 L chain | | Authors: | Wang, Q.H, Wu, Y, Gao, F, Qi, J.X, Gao, G.F. | | Deposit date: | 2018-09-13 | | Release date: | 2019-04-10 | | Last modified: | 2019-07-10 | | Method: | X-RAY DIFFRACTION (2.409 Å) | | Cite: | Neutralization mechanism of human monoclonal antibodies against Rift Valley fever virus.
Nat Microbiol, 4, 2019
|
|
5O44
 
 | | Crystal structure of unbranched mixed tri-Ubiquitin chain containing K48 and K63 linkages. | | Descriptor: | MAGNESIUM ION, Polyubiquitin-B, SULFATE ION, ... | | Authors: | Padala, P, Isupov, M.N, Wiener, R. | | Deposit date: | 2017-05-26 | | Release date: | 2017-11-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | The Crystal Structure and Conformations of an Unbranched Mixed Tri-Ubiquitin Chain Containing K48 and K63 Linkages.
J. Mol. Biol., 429, 2017
|
|
8GPU
 
 | | YFV_E_YD6Fab_prefusion | | Descriptor: | Envelope protein, YD6Fab_H, YD6Fab_L | | Authors: | Li, Y, Wu, L, Qi, J, Yan, J, Gao, G.F. | | Deposit date: | 2022-08-27 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | A neutralizing-protective supersite of human monoclonal antibodies for yellow fever virus.
Innovation (N Y), 3, 2022
|
|
8GPT
 
 | | YFV_E_YD6scFv_postfusion | | Descriptor: | Envelope protein, YD6_VH, YD6_VL | | Authors: | Li, Y, Wu, L, Qi, J, Yan, J, Gao, G.F. | | Deposit date: | 2022-08-27 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | A neutralizing-protective supersite of human monoclonal antibodies for yellow fever virus.
Innovation (N Y), 3, 2022
|
|
7ZSD
 
 | | cryo-EM structure of omicron spike in complex with de novo designed binder, local | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, de novo designed binder | | Authors: | Pablo, G, Sarah, W, Alexandra, V.H, Anthony, M, Andreas, S, Zander, H, Dongchun, N, Shuguang, T, Freyr, S, Casper, G, Priscilla, T, Alexandra, T, Stephane, R, Sandrine, G, Jane, M, Aaron, P, Zepeng, X, Yan, C, Pu, H, George, G, Elisa, O, Beat, F, Didier, T, Henning, S, Michael, B, Bruno, E.C. | | Deposit date: | 2022-05-06 | | Release date: | 2023-03-01 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | De novo design of protein interactions with learned surface fingerprints.
Nature, 617, 2023
|
|
7ZSS
 
 | | cryo-EM structure of D614 spike in complex with de novo designed binder | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Pablo, G, Sarah, W, Alexandra, V.H, Anthony, M, Andreas, S, Zander, H, Dongchun, N, Shuguang, T, Freyr, S, Casper, G, Priscilla, T, Alexandra, T, Stephane, R, Sandrine, G, Jane, M, Aaron, P, Zepeng, X, Yan, C, Pu, H, George, G, Elisa, O, Beat, F, Didier, T, Henning, S, Michael, B, Bruno, E.C. | | Deposit date: | 2022-05-08 | | Release date: | 2023-03-01 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | De novo design of protein interactions with learned surface fingerprints.
Nature, 617, 2023
|
|
8HWT
 
 | |
8HWS
 
 | | The complex structure of Omicron BA.4 RBD with BD604, S309, and S304 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, BD-604 Fab Heavy chain, BD-604 Fab Light chain, ... | | Authors: | He, Q.W, Xu, Z.P, Xie, Y.F. | | Deposit date: | 2023-01-02 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (2.36 Å) | | Cite: | An updated atlas of antibody evasion by SARS-CoV-2 Omicron sub-variants including BQ.1.1 and XBB.
Cell Rep Med, 4, 2023
|
|
7ZRV
 
 | | cryo-EM structure of omicron spike in complex with de novo designed binder, full map | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Envelope glycoprotein, ... | | Authors: | Pablo, G, Sarah, W, Alexandra, V.H, Anthony, M, Andreas, S, Zander, H, Dongchun, N, Shuguang, T, Freyr, S, Casper, G, Priscilla, T, Alexandra, T, Stephane, R, Sandrine, G, Jane, M, Aaron, P, Zepeng, X, Yan, C, Pu, H, George, G, Elisa, O, Beat, F, Didier, T, Henning, S, Michael, B, Bruno, E.C. | | Deposit date: | 2022-05-05 | | Release date: | 2023-03-08 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | De novo design of protein interactions with learned surface fingerprints.
Nature, 617, 2023
|
|
