2KHO
 
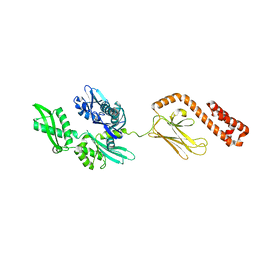 | |
1K46
 
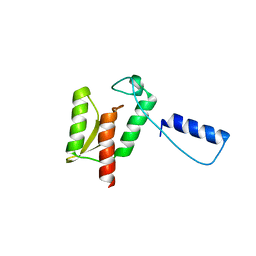 | | Crystal Structure of the Type III Secretory Domain of Yersinia YopH Reveals a Domain-Swapped Dimer | | Descriptor: | PROTEIN-TYROSINE PHOSPHATASE YOPH | | Authors: | Smith, C.L, Khandelwal, P, Keliikuli, K, Zuiderweg, E.R.P, Saper, M.A. | | Deposit date: | 2001-10-05 | | Release date: | 2001-11-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the type III secretion and substrate-binding domain of Yersinia YopH phosphatase.
Mol.Microbiol., 42, 2001
|
|
1UMT
 
 | | Stromelysin-1 catalytic domain with hydrophobic inhibitor bound, ph 7.0, 32oc, 20 mm cacl2, 15% acetonitrile; nmr average of 20 structures minimized with restraints | | Descriptor: | CALCIUM ION, N-{(2R)-2-[2-(hydroxyamino)-2-oxoethyl]-4-methylpentanoyl}-L-leucyl-L-phenylalaninamide, STROMELYSIN-1, ... | | Authors: | Van Doren, S.R, Kurochkin, A.V, Hu, W, Zuiderweg, E.R.P. | | Deposit date: | 1995-10-31 | | Release date: | 1996-03-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the catalytic domain of human stromelysin complexed with a hydrophobic inhibitor.
Protein Sci., 4, 1995
|
|
1UMS
 
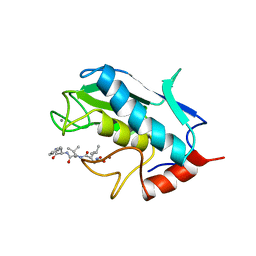 | | STROMELYSIN-1 CATALYTIC DOMAIN WITH HYDROPHOBIC INHIBITOR BOUND, PH 7.0, 32OC, 20 MM CACL2, 15% ACETONITRILE; NMR ENSEMBLE OF 20 STRUCTURES | | Descriptor: | CALCIUM ION, N-{(2R)-2-[2-(hydroxyamino)-2-oxoethyl]-4-methylpentanoyl}-L-leucyl-L-phenylalaninamide, STROMELYSIN-1, ... | | Authors: | Van Doren, S.R, Kurochkin, A.V, Hu, W, Zuiderweg, E.R.P. | | Deposit date: | 1995-10-31 | | Release date: | 1996-03-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the catalytic domain of human stromelysin complexed with a hydrophobic inhibitor.
Protein Sci., 4, 1995
|
|
1Q5L
 
 | | NMR structure of the substrate binding domain of DnaK bound to the peptide NRLLLTG | | Descriptor: | Chaperone protein dnaK, peptide NRLLLTG | | Authors: | Stevens, S.Y, Cai, S, Pellecchia, M, Zuiderweg, E.R. | | Deposit date: | 2003-08-08 | | Release date: | 2003-11-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the bacterial HSP70 chaperone protein domain DnaK(393-507) in complex with the peptide NRLLLTG.
Protein Sci., 12, 2003
|
|
2BPR
 
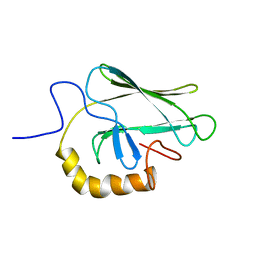 | | NMR STRUCTURE OF THE SUBSTRATE BINDING DOMAIN OF DNAK, 25 STRUCTURES | | Descriptor: | DNAK | | Authors: | Wang, H, Kurochkin, A.V, Pang, Y, Hu, W, Flynn, G.C, Zuiderweg, E.R.P. | | Deposit date: | 1998-08-11 | | Release date: | 1999-03-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the 21 kDa chaperone protein DnaK substrate binding domain: a preview of chaperone-protein interaction.
Biochemistry, 37, 1998
|
|
1BPR
 
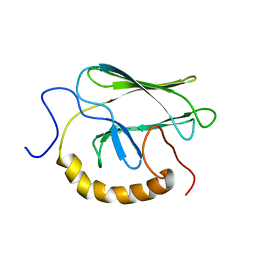 | | NMR STRUCTURE OF THE SUBSTRATE BINDING DOMAIN OF DNAK, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DNAK | | Authors: | Wang, H, Kurochkin, A.V, Pang, Y, Hu, W, Flynn, G.C, Zuiderweg, E.R.P. | | Deposit date: | 1998-08-11 | | Release date: | 1999-03-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the 21 kDa chaperone protein DnaK substrate binding domain: a preview of chaperone-protein interaction.
Biochemistry, 37, 1998
|
|
1CKR
 
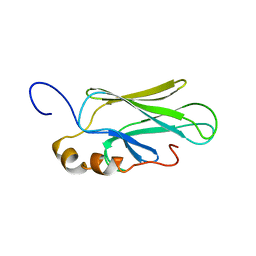 | | HIGH RESOLUTION SOLUTION STRUCTURE OF THE HEAT SHOCK COGNATE-70 KD SUBSTRATE BINDING DOMAIN OBTAINED BY MULTIDIMENSIONAL NMR TECHNIQUES | | Descriptor: | HEAT SHOCK SUBSTRATE BINDING DOMAIN OF HSC-70 | | Authors: | Morshauser, R.C, Hu, W, Wang, H, Pang, Y, Flynn, G.C, Zuiderweg, E.R.P. | | Deposit date: | 1999-04-22 | | Release date: | 1999-04-30 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution structure of the 18 kDa substrate-binding domain of the mammalian chaperone protein Hsc70.
J.Mol.Biol., 289, 1999
|
|
1DG4
 
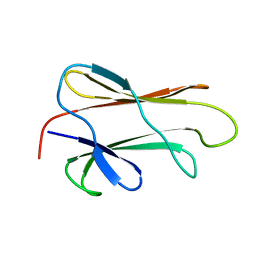 | | NMR STRUCTURE OF THE SUBSTRATE BINDING DOMAIN OF DNAK IN THE APO FORM | | Descriptor: | DNAK | | Authors: | Pellecchia, M, Montgomery, D.L, Stevens, S.Y, Van der Kooi, C.W, Feng, H, Gierasch, L.M, Zuiderweg, E.R.P. | | Deposit date: | 1999-11-23 | | Release date: | 1999-12-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural insights into substrate binding by the molecular chaperone DnaK.
Nat.Struct.Biol., 7, 2000
|
|
7HSC
 
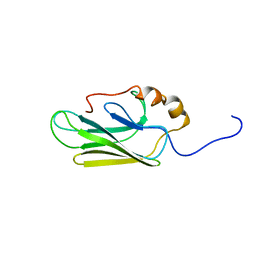 | | HIGH RESOLUTION SOLUTION STRUCTURE OF THE HEAT SHOCK COGNATE-70 KD SUBSTRATE BINDING DOMAIN OBTAINED BY MULTIDIMENSIONAL NMR TECHNIQUES | | Descriptor: | PROTEIN (HEAT SHOCK COGNATE 70 KD PROTEIN 1) | | Authors: | Morshauser, R.C, Hu, W, Wang, H, Pang, Y, Flynn, G.C, Zuiderweg, E.R.P. | | Deposit date: | 1999-05-03 | | Release date: | 1999-05-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution structure of the 18 kDa substrate-binding domain of the mammalian chaperone protein Hsc70.
J.Mol.Biol., 289, 1999
|
|
2MTQ
 
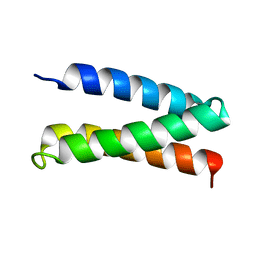 | | Solution Structure of a De Novo Designed Peptide that Sequesters Toxic Heavy Metals | | Descriptor: | Designed Peptide | | Authors: | Plegaria, J.S, Zuiderweg, E.R, Stemmler, T.L, Pecoraro, V.L. | | Deposit date: | 2014-08-28 | | Release date: | 2015-04-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Apoprotein Structure and Metal Binding Characterization of a de Novo Designed Peptide, alpha 3DIV, that Sequesters Toxic Heavy Metals.
Biochemistry, 54, 2015
|
|
1HOF
 
 | |
1HO9
 
 | |
1HOD
 
 | |
1HLL
 
 | |
1M0V
 
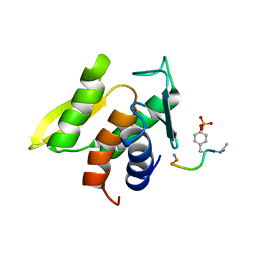 | | NMR STRUCTURE OF THE TYPE III SECRETORY DOMAIN OF YERSINIA YOPH COMPLEXED WITH THE SKAP-HOM PHOSPHO-PEPTIDE N-acetyl-DEpYDDPF-NH2 | | Descriptor: | PROTEIN-TYROSINE PHOSPHATASE YOPH, SKAP55 homologue | | Authors: | Khandelwal, P, Keliikuli, K, Smith, C.L, Saper, M.A, Zuiderweg, E.R.P. | | Deposit date: | 2002-06-14 | | Release date: | 2002-07-24 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure and phosphopeptide binding to the N-terminal domain of Yersinia YopH: comparison with a crystal structure
Biochemistry, 41, 2002
|
|
1LQC
 
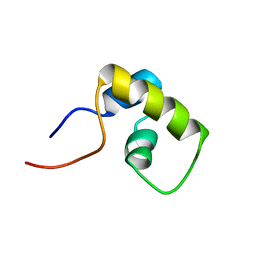 | | LAC REPRESSOR HEADPIECE (RESIDUES 1-56), NMR, 32 STRUCTURES | | Descriptor: | LAC REPRESSOR | | Authors: | Slijper, M, Bonvin, A.M.J.J, Boelens, R, Kaptein, R. | | Deposit date: | 1996-08-13 | | Release date: | 1997-02-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Refined structure of lac repressor headpiece (1-56) determined by relaxation matrix calculations from 2D and 3D NOE data: change of tertiary structure upon binding to the lac operator.
J.Mol.Biol., 259, 1996
|
|
5IOS
 
 | |
5IOT
 
 | |
5IOQ
 
 | | Flavin-dependent thymidylate synthase in complex with FAD and deoxyuridine | | Descriptor: | 2'-DEOXYURIDINE, FLAVIN-ADENINE DINUCLEOTIDE, TRIETHYLENE GLYCOL, ... | | Authors: | Bernard, S.M, Stull, F.W, Smith, J.L. | | Deposit date: | 2016-03-08 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Deprotonations in the Reaction of Flavin-Dependent Thymidylate Synthase.
Biochemistry, 55, 2016
|
|
5IOR
 
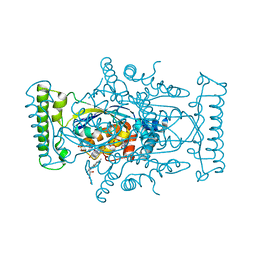 | | Flavin-dependent thymidylate synthase in complex with FAD and 2'-deoxyuridine-5'-monosulfate | | Descriptor: | 2'-deoxy-5'-O-sulfouridine, FLAVIN-ADENINE DINUCLEOTIDE, RIBOFLAVIN, ... | | Authors: | Bernard, S.M, Stull, F.W, Smith, J.L. | | Deposit date: | 2016-03-08 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Deprotonations in the Reaction of Flavin-Dependent Thymidylate Synthase.
Biochemistry, 55, 2016
|
|
