7Y6B
 
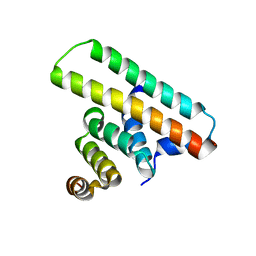 | | Crystal structure of the EscE/EsaH complex | | Descriptor: | EscE/YscE/SsaE family type III secretion system needle protein co-chaperone, EscG/YscG/SsaH family type III secretion system needle protein co-chaperone | | Authors: | Zeng, Z.X, Xiao, S.B, Liao, L.J, Zhou, Y.Z. | | Deposit date: | 2022-06-18 | | Release date: | 2022-07-20 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Secreted in a Type III Secretion System-Dependent Manner, EsaH and EscE Are the Cochaperones of the T3SS Needle Protein EsaG of Edwardsiella piscicida.
Mbio, 13, 2022
|
|
7Y6C
 
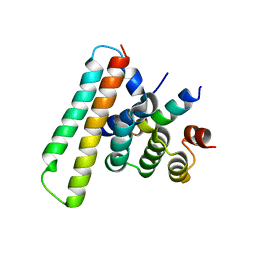 | | Crystal structure of the EscE/EsaG/EsaH complex | | Descriptor: | EscE/YscE/SsaE family type III secretion system needle protein co-chaperone, EscG/YscG/SsaH family type III secretion system needle protein co-chaperone | | Authors: | Zeng, Z.X, Xiao, S.B, Liao, L.J, Zhou, Y.Z. | | Deposit date: | 2022-06-18 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Secreted in a Type III Secretion System-Dependent Manner, EsaH and EscE Are the Cochaperones of the T3SS Needle Protein EsaG of Edwardsiella piscicida.
Mbio, 13, 2022
|
|
7XIN
 
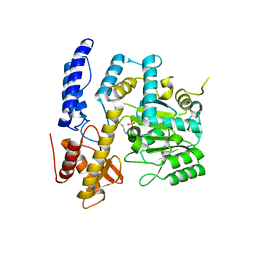 | | Crystal structure of DODC from Pseudomonas | | Descriptor: | DOPA decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Li, X, Zhou, Y.L, Liao, L.J, Liu, X.K, Liu, B, Guo, Y, Feng, Z, Sun, D.Y, Zeng, Z.X. | | Deposit date: | 2022-04-13 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of DODC from Pseudomonas
To Be Published
|
|
7XIO
 
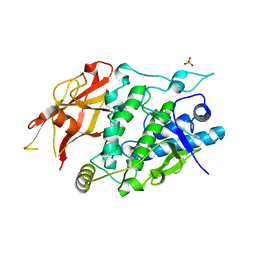 | | Crystal structure of TYR from Ralstonia | | Descriptor: | PHOSPHATE ION, Polyphenol oxidase | | Authors: | Sun, D.Y, Cui, P.P, Liao, L.J, Liu, X.K, Liu, B, Guo, Y, Feng, Z, Zhang, J, Li, X, Zeng, Z.X. | | Deposit date: | 2022-04-13 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Crystal structure of TYR from Ralstonia
To Be Published
|
|
6IV7
 
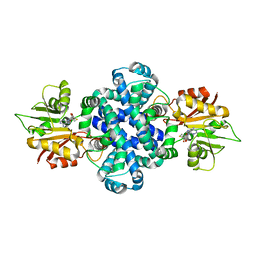 | | The crystal structure of a SAM-dependent enzyme from aspergillus flavus | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, methyltransferase lepI | | Authors: | Zhou, Y.Z, Peng, T, Liao, L.J, Liu, X.K, Zhao, Y.C, Guo, Y, Zeng, Z.X. | | Deposit date: | 2018-12-02 | | Release date: | 2019-02-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.937 Å) | | Cite: | The crystal structure of A SAM-Dependent enzyme from Aspergillus flavus
To Be Published
|
|
6K5S
 
 | |
6K5U
 
 | |
3L82
 
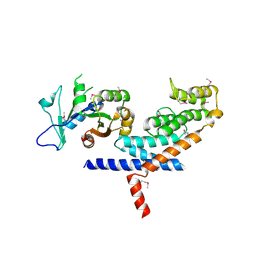 | | X-ray Crystal structure of TRF1 and Fbx4 complex | | Descriptor: | F-box only protein 4, Telomeric repeat-binding factor 1 | | Authors: | Zeng, Z.X, Wang, W, Yang, Y.T, Chen, Y, Yang, X.M, Diehl, J.A, Liu, X.D, Lei, M. | | Deposit date: | 2009-12-29 | | Release date: | 2010-03-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of Selective Ubiquitination of TRF1 by SCF(Fbx4)
Dev.Cell, 18, 2010
|
|
6IWT
 
 | |
6K5M
 
 | | The crystal structure of a serotonin N-acetyltransferase from Oryza Sativa (Rice) | | Descriptor: | Serotonin N-acetyltransferase 1, chloroplastic | | Authors: | Zhou, Y.Z, Liao, L.J, Liu, X.K, Guo, Y, Zhao, Y.C, Zeng, Z.X. | | Deposit date: | 2019-05-29 | | Release date: | 2020-06-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.793 Å) | | Cite: | Structural and Molecular Dynamics Analysis of Plant Serotonin N-Acetyltransferase Reveal an Acid/Base-Assisted Catalysis in Melatonin Biosynthesis.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6KHO
 
 | | Crystal structure of Oryza sativa TDC with PLP | | Descriptor: | ACETATE ION, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhou, Y.Z, Liao, L.J, Liu, X.K, Guo, Y, Zhao, Y.C, Zeng, Z.X. | | Deposit date: | 2019-07-16 | | Release date: | 2020-07-15 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.972 Å) | | Cite: | Crystal structure ofOryza sativaTDC reveals the substrate specificity for TDC-mediated melatonin biosynthesis.
J Adv Res, 24, 2020
|
|
6KHN
 
 | | Crystal structure of Oryza sativa TDC with PLP and SEROTONIN | | Descriptor: | ACETATE ION, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhou, Y.Z, Liao, L.J, Liu, X.K, Guo, Y, Zhao, Y.C, Zeng, Z.X. | | Deposit date: | 2019-07-16 | | Release date: | 2020-07-15 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.293 Å) | | Cite: | Crystal structure ofOryza sativaTDC reveals the substrate specificity for TDC-mediated melatonin biosynthesis.
J Adv Res, 24, 2020
|
|
6KHP
 
 | | Crystal structure of Oryza sativa TDC with PLP and tryptamine | | Descriptor: | 2-(1H-INDOL-3-YL)ETHANAMINE, ACETATE ION, CALCIUM ION, ... | | Authors: | Zhou, Y.Z, Liao, L.J, Liu, X.K, Guo, Y, Zhao, Y.C, Zeng, Z.X. | | Deposit date: | 2019-07-16 | | Release date: | 2020-07-15 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Crystal structure ofOryza sativaTDC reveals the substrate specificity for TDC-mediated melatonin biosynthesis.
J Adv Res, 24, 2020
|
|
