5G0A
 
 | | The crystal structure of a S-selective transaminase from Bacillus megaterium | | Descriptor: | DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | van Oosterwijk, N, Willies, S, Hekelaar, J, Terwisscha van Scheltinga, A.C, Turner, N.J, Dijkstra, B.W. | | Deposit date: | 2016-03-17 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of Substrate Range and Enantioselectivity of Two S-Selective Omega- Transaminases
Biochemistry, 55, 2016
|
|
5G09
 
 | | The crystal structure of a S-selective transaminase from Bacillus megaterium bound with R-alpha-methylbenzylamine | | Descriptor: | DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, TETRAETHYLENE GLYCOL, ... | | Authors: | van Oosterwijk, N, Willies, S, Hekelaar, J, Terwisscha van Scheltinga, A.C, Turner, N.J, Dijkstra, B.W. | | Deposit date: | 2016-03-17 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis of Substrate Range and Enantioselectivity of Two S-Selective Omega- Transaminases
Biochemistry, 55, 2016
|
|
5G2Q
 
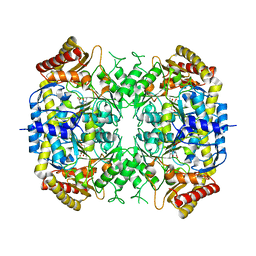 | | The crystal structure of a S-selective transaminase from Arthrobacter sp. with alanine bound | | Descriptor: | 2-[(3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL)-AMINO]-PROPIONIC ACID, TRANSAMINASE | | Authors: | van Oosterwijk, N, Willies, S, Hekelaar, J, Terwisscha van Scheltinga, A.C, Turner, N.J, Dijkstra, B.W. | | Deposit date: | 2016-04-12 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of Substrate Range and Enantioselectivity of Two S-Selective Omega- Transaminases
Biochemistry, 55, 2016
|
|
5G2P
 
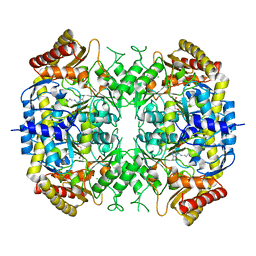 | | The crystal structure of a S-selective transaminase from Arthrobacter sp. | | Descriptor: | CHLORIDE ION, PYRIDOXAL-5'-PHOSPHATE, TRANSAMINASE | | Authors: | van Oosterwijk, N, Willies, S, Hekelaar, J, Terwisscha van Scheltinga, A.C, Turner, N.J, Dijkstra, B.W. | | Deposit date: | 2016-04-12 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural Basis of Substrate Range and Enantioselectivity of Two S-Selective Omega- Transaminases
Biochemistry, 55, 2016
|
|
7ZX1
 
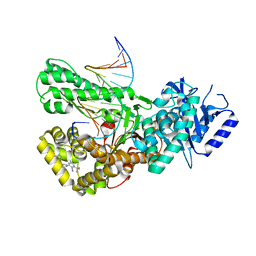 | | Crystal structure of Pol theta polymerase domain in complex with compound 22 | | Descriptor: | (2~{S},3~{R})-1-[3-cyano-6-methyl-4-(trifluoromethyl)pyridin-2-yl]-~{N}-methyl-~{N}-(3-methylphenyl)-3-oxidanyl-pyrrolidine-2-carboxamide, 2'-3'-DIDEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA (5'-D(*GP*CP*GP*GP*CP*TP*GP*TP*CP*AP*TP*TP*(DDG))-3'), ... | | Authors: | Krajewski, W.W, Turnbull, A.P, Willis, S, Charles, M, Stockley, M, Heald, R.A. | | Deposit date: | 2022-05-19 | | Release date: | 2022-10-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.829 Å) | | Cite: | Discovery, Characterization, and Structure-Based Optimization of Small-Molecule In Vitro and In Vivo Probes for Human DNA Polymerase Theta.
J.Med.Chem., 65, 2022
|
|
7ZX0
 
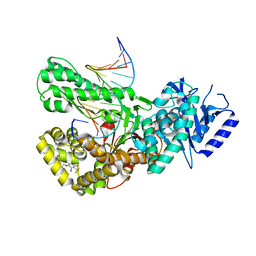 | | Crystal structure of Pol theta polymerase domain in complex with compound 5 | | Descriptor: | 2'-3'-DIDEOXYGUANOSINE-5'-TRIPHOSPHATE, 2-[5-bromanyl-3-cyano-6-methyl-4-(trifluoromethyl)pyridin-2-yl]oxy-~{N}-ethyl-~{N}-(3-methylphenyl)ethanamide, DNA (5'-D(*GP*CP*GP*GP*CP*TP*GP*TP*CP*AP*TP*TP*(DDG))-3'), ... | | Authors: | Krajewski, W.W, Turnbull, A.P, Willis, S, Charles, M, Stockley, M, Heald, R.A. | | Deposit date: | 2022-05-19 | | Release date: | 2022-10-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Discovery, Characterization, and Structure-Based Optimization of Small-Molecule In Vitro and In Vivo Probes for Human DNA Polymerase Theta.
J.Med.Chem., 65, 2022
|
|
7ZUS
 
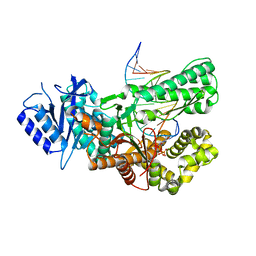 | | Crystal structure of ternary complex of Pol theta polymerase domain | | Descriptor: | 2'-3'-DIDEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA (5'-D(*GP*CP*GP*GP*CP*TP*GP*TP*CP*AP*TP*TP*(DDG))-3'), DNA (5'-D(P*TP*TP*CP*CP*AP*AP*TP*GP*AP*CP*AP*GP*CP*CP*GP*C)-3'), ... | | Authors: | Krajewski, W.W, Turnbull, A.P, Willis, S, Charles, M, Stockley, M, Heald, R.A. | | Deposit date: | 2022-05-13 | | Release date: | 2022-10-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Discovery, Characterization, and Structure-Based Optimization of Small-Molecule In Vitro and In Vivo Probes for Human DNA Polymerase Theta.
J.Med.Chem., 65, 2022
|
|
1B3J
 
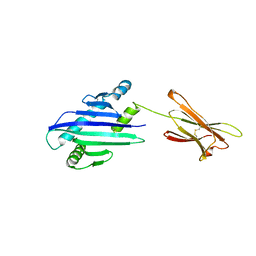 | | STRUCTURE OF THE MHC CLASS I HOMOLOG MIC-A, A GAMMADELTA T CELL LIGAND | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MHC CLASS I HOMOLOG MIC-A | | Authors: | Li, P, Willie, S, Bauer, S, Morris, D, Spies, T, Strong, R. | | Deposit date: | 1998-12-11 | | Release date: | 1999-07-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the MHC class I homolog MIC-A, a gammadelta T cell ligand.
Immunity, 10, 1999
|
|
3ZDN
 
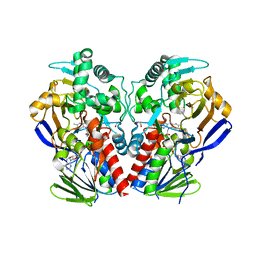 | | D11-C mutant of monoamine oxidase from Aspergillus niger | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, MONOAMINE OXIDASE N | | Authors: | Frank, A, Ghislieri, D, Willies, S, Turner, N.J, Grogan, G. | | Deposit date: | 2012-11-29 | | Release date: | 2013-10-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Engineering an Enantioselective Amine Oxidase for the Synthesis of Pharmaceutical Building Blocks and Alkaloid Natural Products.
J.Am.Chem.Soc., 135, 2013
|
|
8P2E
 
 | | Homotypic interacting B1 fab bound to Chondroitin Sulfate A | | Descriptor: | B1 fab heavy, B1 fab light | | Authors: | Raghavan, S.S.R, Dagil, R, Wang, K.T, Salanti, A. | | Deposit date: | 2023-05-16 | | Release date: | 2024-06-05 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Tumor-agnostic cancer therapy using antibodies targeting oncofetal chondroitin sulfate.
Nat Commun, 15, 2024
|
|
4YWR
 
 | |
4YL5
 
 | |
6GOL
 
 | |
