3L41
 
 | | Crystal Structure of S. pombe Brc1 BRCT5-BRCT6 domains in complex with phosphorylated H2A | | Descriptor: | BRCT-containing protein 1, phosphorylated H2A tail | | Authors: | Williams, R.S, Williams, J.S, Guenther, G, Tainer, J.A. | | Deposit date: | 2009-12-18 | | Release date: | 2010-02-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | gammaH2A binds Brc1 to maintain genome integrity during S-phase.
Embo J., 29, 2010
|
|
3L40
 
 | |
7N3Z
 
 | | Crystal Structure of Saccharomyces cerevisiae Apn2 Catalytic Domain | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA-(apurinic or apyrimidinic site) endonuclease 2, ... | | Authors: | Wojtaszek, J.L, Wallace, B.D, Williams, R.S. | | Deposit date: | 2021-06-02 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Molecular basis for processing of topoisomerase 1-triggered DNA damage by Apn2/APE2.
Cell Rep, 41, 2022
|
|
7N3Y
 
 | | Crystal Structure of Saccharomyces cerevisiae Apn2 Catalytic Domain E59Q/D222N Mutant in Complex with DNA | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CITRIC ACID, ... | | Authors: | Wojtaszek, J.L, Krahn, J, Wallace, B.D, Williams, R.S. | | Deposit date: | 2021-06-02 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Molecular basis for processing of topoisomerase 1-triggered DNA damage by Apn2/APE2.
Cell Rep, 41, 2022
|
|
7KR3
 
 | |
7KR4
 
 | |
3SZQ
 
 | | Structure of an S. pombe APTX/DNA/AMP/Zn complex | | Descriptor: | 5'-D(*CP*CP*CP*TP*G)-3', 5'-D(*TP*AP*TP*CP*GP*GP*AP*AP*TP*CP*AP*GP*GP*G)-3', ADENOSINE MONOPHOSPHATE, ... | | Authors: | Tumbale, P, Krahn, J, Williams, R.S. | | Deposit date: | 2011-07-19 | | Release date: | 2011-10-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.353 Å) | | Cite: | Structure of an aprataxin-DNA complex with insights into AOA1 neurodegenerative disease.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3QKU
 
 | | Mre11 Rad50 binding domain in complex with Rad50 and AMP-PNP | | Descriptor: | DNA double-strand break repair protein mre11, DNA double-strand break repair rad50 ATPase, MAGNESIUM ION, ... | | Authors: | Williams, G.J, Williams, R.S, Arvai, A, Moncalian, G, Tainer, J.A. | | Deposit date: | 2011-02-01 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | ABC ATPase signature helices in Rad50 link nucleotide state to Mre11 interface for DNA repair.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3QKS
 
 | | Mre11 Rad50 binding domain bound to Rad50 | | Descriptor: | DNA double-strand break repair protein mre11, DNA double-strand break repair rad50 ATPase | | Authors: | Williams, G.J, Williams, R.S, Arvai, A, Moncalian, G, Tainer, J.A. | | Deposit date: | 2011-02-01 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | ABC ATPase signature helices in Rad50 link nucleotide state to Mre11 interface for DNA repair.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3QKT
 
 | | Rad50 ABC-ATPase with adjacent coiled-coil region in complex with AMP-PNP | | Descriptor: | DNA double-strand break repair rad50 ATPase, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Williams, G.J, Williams, R.S, Arvai, A, Moncalian, G, Tainer, J.A. | | Deposit date: | 2011-02-01 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | ABC ATPase signature helices in Rad50 link nucleotide state to Mre11 interface for DNA repair.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3QKR
 
 | | Mre11 Rad50 binding domain bound to Rad50 | | Descriptor: | DNA double-strand break repair protein mre11, DNA double-strand break repair rad50 ATPase, PHOSPHATE ION | | Authors: | Williams, G.J, Williams, R.S, Arvai, A, Moncalian, G, Tainer, J.A. | | Deposit date: | 2011-02-01 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | ABC ATPase signature helices in Rad50 link nucleotide state to Mre11 interface for DNA repair.
Nat.Struct.Mol.Biol., 18, 2011
|
|
4X01
 
 | | S. pombe Ctp1 tetramerization domain | | Descriptor: | 1,2-ETHANEDIOL, DNA binding ctp1 | | Authors: | Andres, S.N, Williams, R.S. | | Deposit date: | 2014-11-20 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Tetrameric Ctp1 coordinates DNA binding and DNA bridging in DNA double-strand-break repair.
Nat.Struct.Mol.Biol., 22, 2015
|
|
3HUE
 
 | |
3HUF
 
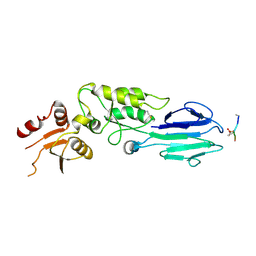 | | Structure of the S. pombe Nbs1-Ctp1 complex | | Descriptor: | DNA repair and telomere maintenance protein nbs1, Double-strand break repair protein ctp1, THIOCYANATE ION | | Authors: | Williams, R.S, Guenther, G, Tainer, J.A. | | Deposit date: | 2009-06-13 | | Release date: | 2009-10-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Nbs1 flexibly tethers Ctp1 and Mre11-Rad50 to coordinate DNA double-strand break processing and repair.
Cell(Cambridge,Mass.), 139, 2009
|
|
4NDG
 
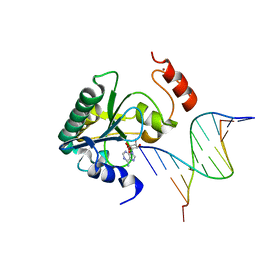 | | Human Aprataxin (Aptx) bound to RNA-DNA and Zn - adenosine vanadate transition state mimic complex | | Descriptor: | 5'-D(*GP*AP*AP*TP*CP*AP*TP*AP*AP*C)-3', 5'-R(P*G)-D(P*TP*TP*AP*TP*GP*AP*TP*TP*C)-3', Aprataxin, ... | | Authors: | Schellenberg, M.J, Tumbale, P.S, Williams, R.S. | | Deposit date: | 2013-10-26 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.541 Å) | | Cite: | Aprataxin resolves adenylated RNA-DNA junctions to maintain genome integrity.
Nature, 506, 2013
|
|
4NDF
 
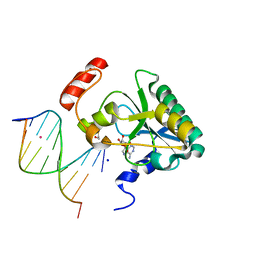 | | Human Aprataxin (Aptx) bound to RNA-DNA, AMP, and Zn - product complex | | Descriptor: | 5'-D(*GP*AP*AP*TP*CP*AP*TP*AP*AP*C)-3', 5'-R(P*G)-D(P*TP*TP*AP*TP*GP*AP*TP*TP*C)-3', ADENOSINE MONOPHOSPHATE, ... | | Authors: | Schellenberg, M.J, Tumbale, P.S, Williams, R.S. | | Deposit date: | 2013-10-26 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.944 Å) | | Cite: | Aprataxin resolves adenylated RNA-DNA junctions to maintain genome integrity.
Nature, 506, 2013
|
|
4NDI
 
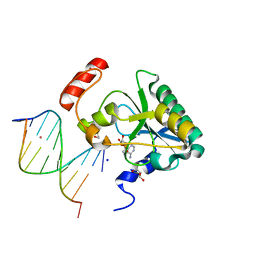 | | Human Aprataxin (Aptx) AOA1 variant K197Q bound to RNA-DNA, AMP, and Zn - product complex | | Descriptor: | 5'-D(*GP*AP*AP*TP*CP*AP*TP*AP*AP*C)-3', 5'-R(P*G)-D(P*TP*TP*AP*TP*GP*AP*TP*TP*C)-3', ADENOSINE MONOPHOSPHATE, ... | | Authors: | Schellenberg, M.J, Tumbale, P.S, Williams, R.S. | | Deposit date: | 2013-10-26 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Aprataxin resolves adenylated RNA-DNA junctions to maintain genome integrity.
Nature, 506, 2013
|
|
4NDH
 
 | | Human Aprataxin (Aptx) bound to DNA, AMP, and Zn - product complex | | Descriptor: | 5'-D(P*GP*TP*TP*CP*TP*AP*GP*AP*AP*C)-3', ADENOSINE MONOPHOSPHATE, Aprataxin, ... | | Authors: | Schellenberg, M.J, Tumbale, P.S, Williams, R.S. | | Deposit date: | 2013-10-26 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Aprataxin resolves adenylated RNA-DNA junctions to maintain genome integrity.
Nature, 506, 2013
|
|
3DSC
 
 | | Crystal structure of P. furiosus Mre11 DNA synaptic complex | | Descriptor: | DNA (5'-D(P*DCP*DAP*DCP*DAP*DAP*DGP*DCP*DTP*DTP*DTP*DTP*DGP*DCP*DTP*DTP*DGP*DTP*DGP*DAP*DC)-3'), DNA double-strand break repair protein mre11 | | Authors: | Williams, R.S, Moncalian, G, Shin, D.S, Tainer, J.A. | | Deposit date: | 2008-07-11 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair.
Cell(Cambridge,Mass.), 135, 2008
|
|
3DSD
 
 | | Crystal structure of P. furiosus Mre11-H85S bound to a branched DNA and manganese | | Descriptor: | DNA (5'-D(*DCP*DGP*DCP*DGP*DCP*DAP*DCP*DAP*DAP*DGP*DCP*DTP*DTP*DTP*DTP*DGP*DCP*DTP*DTP*DGP*DTP*DGP*DGP*DAP*DTP*DA)-3'), DNA double-strand break repair protein mre11, MANGANESE (II) ION | | Authors: | Williams, R.S, Moiani, D, Tainer, J.A. | | Deposit date: | 2008-07-11 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair.
Cell(Cambridge,Mass.), 135, 2008
|
|
1ZP5
 
 | | Crystal structure of the complex between MMP-8 and a N-hydroxyurea inhibitor | | Descriptor: | CALCIUM ION, N-{2-[(4'-CYANO-1,1'-BIPHENYL-4-YL)OXY]ETHYL}-N'-HYDROXY-N-METHYLUREA, Neutrophil collagenase, ... | | Authors: | Campestre, C, Agamennone, M, Tortorella, P, Preziuso, S, Biasone, A, Gavuzzo, E, Pochetti, G, Mazza, F, Tschesche, H, Gallina, C. | | Deposit date: | 2005-05-16 | | Release date: | 2005-12-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | N-Hydroxyurea as zinc binding group in matrix metalloproteinase inhibition: Mode of binding in a complex with MMP-8.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
