2KUS
 
 | |
2L3I
 
 | | Oxki4a, spider derived antimicrobial peptide | | Descriptor: | AOXKI4A, antimicrobial peptide in spider venom | | Authors: | Vassilevski, A.A, Dubovskii, P.V, Samsonova, O.V, Egorova, N.S, Kozlov, S.A, Feofanov, A.V, Arseniev, A.S, Grishin, E.V. | | Deposit date: | 2010-09-14 | | Release date: | 2011-09-14 | | Last modified: | 2019-05-15 | | Method: | SOLUTION NMR | | Cite: | Novel lynx spider toxin shares common molecular architecture with defense peptides from frog skin.
Febs J., 278, 2011
|
|
6THI
 
 | | Solution structure of MeuNaTxalpha-1 toxin from Mesobuthus Eupeus | | Descriptor: | Sodium channel neurotoxin MeuNaTxalpha-1 | | Authors: | Mineev, K.S, Kuzmenkov, A.I, Khusainov, G.A, Arseniev, A.S, Vassilevski, A.A. | | Deposit date: | 2019-11-20 | | Release date: | 2020-12-02 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Structure of MeuNaTx alpha-1 toxin from scorpion venom highlights the importance of the nest motif.
Proteins, 2021
|
|
7QXJ
 
 | |
6O9G
 
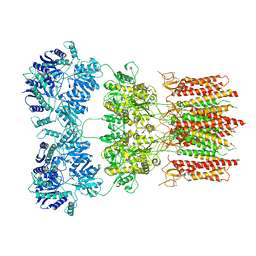 | | Open state GluA2 in complex with STZ and blocked by AgTx-636, after micelle signal subtraction | | Descriptor: | CYCLOTHIAZIDE, GLUTAMIC ACID, Glutamate receptor 2,Voltage-dependent calcium channel gamma-2 subunit, ... | | Authors: | Twomey, E.C, Yelshanskaya, M.V, Vassilevski, A.A, Sobolevsky, A.I. | | Deposit date: | 2019-03-13 | | Release date: | 2019-03-20 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Mechanisms of Channel Block in Calcium-Permeable AMPA Receptors.
Neuron, 99, 2018
|
|
5MOU
 
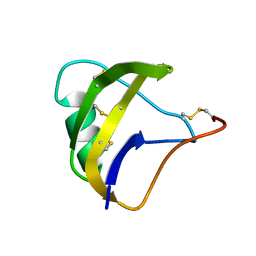 | | NMR spatial structure of scorpion alpha-like toxin BeM9 | | Descriptor: | Alpha-mammal toxin BeM9 | | Authors: | Mineev, K.S, Kuldushev, N.A, Berkut, A.A, Grishin, E.V, Vassilevski, A.A, Arseniev, A.S. | | Deposit date: | 2016-12-14 | | Release date: | 2018-01-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Refined structure of BeM9 reveals arginine hand, an overlooked structural motif in scorpion toxins affecting sodium channels.
Proteins, 86, 2018
|
|
5LM0
 
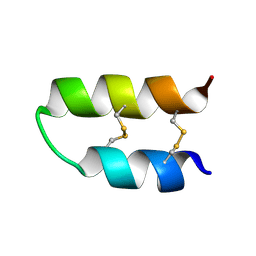 | | NMR spatial structure of Tk-hefu peptide | | Descriptor: | L-2 | | Authors: | Mineev, K.S, Berkut, A.A, Novikova, E.V, Oparin, P.B, Grishin, E.V, Arseniev, A.S, Vassilevski, A.A. | | Deposit date: | 2016-07-28 | | Release date: | 2017-08-16 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Protein Surface Topography as a tool to enhance the selective activity of a potassium channel blocker.
J.Biol.Chem., 2019
|
|
8BO0
 
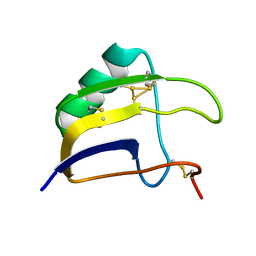 | | Solution structure of Lqq4 toxin from Leiurus quinquestriatus quinquestriatus | | Descriptor: | Alpha-toxin Lqq4 | | Authors: | Mineev, K.S, Motov, V.V, Vassilevski, A.A, Chernykh, M.A, Kuzmenkov, A.I. | | Deposit date: | 2022-11-14 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | SOLUTION NMR | | Cite: | A scorpion toxin affecting sodium channels shows double cis-trans isomerism.
Febs Lett., 597, 2023
|
|
2LB7
 
 | | Hevein-type Antifungal Peptide with a Unique 10-Cysteine Motif | | Descriptor: | Antimicrobial peptide 1a | | Authors: | Balashova, T.A, Vassilevski, A.A, Odintsova, T.I, Grishin, E.V, Egorov, T.A, Arseniev, A.S. | | Deposit date: | 2011-03-23 | | Release date: | 2011-04-13 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a defense peptide from wheat with a 10-cysteine motif.
Biochem.Biophys.Res.Commun., 411, 2011
|
|
8CM3
 
 | |
6DM0
 
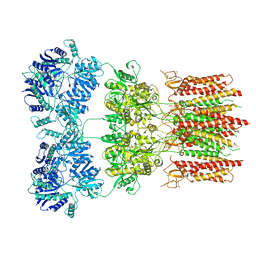 | | Open state GluA2 in complex with STZ and blocked by IEM-1460, after micelle signal subtraction | | Descriptor: | CYCLOTHIAZIDE, GLUTAMIC ACID, Glutamate receptor 2,Voltage-dependent calcium channel gamma-2 subunit, ... | | Authors: | Twomey, E.C, Yelshanskaya, M.V, Vassilevski, A.A, Sobolevsky, A.I. | | Deposit date: | 2018-06-04 | | Release date: | 2018-08-22 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Mechanisms of Channel Block in Calcium-Permeable AMPA Receptors.
Neuron, 99, 2018
|
|
6DM1
 
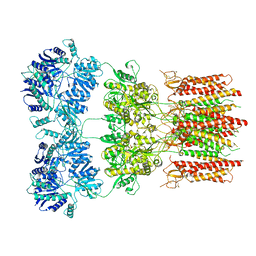 | | Open state GluA2 in complex with STZ and blocked by NASPM, after micelle signal subtraction | | Descriptor: | CYCLOTHIAZIDE, GLUTAMIC ACID, Glutamate receptor 2,Voltage-dependent calcium channel gamma-2 subunit, ... | | Authors: | Twomey, E.C, Yelshanskaya, M.V, Vassilevski, A.A, Sobolevsky, A.I. | | Deposit date: | 2018-06-04 | | Release date: | 2018-08-22 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Mechanisms of Channel Block in Calcium-Permeable AMPA Receptors.
Neuron, 99, 2018
|
|
6DLZ
 
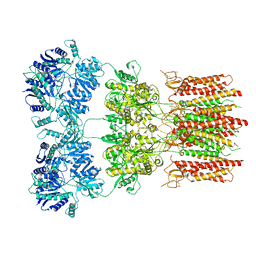 | | Open state GluA2 in complex with STZ after micelle signal subtraction | | Descriptor: | CYCLOTHIAZIDE, GLUTAMIC ACID, Glutamate receptor 2,Voltage-dependent calcium channel gamma-2 subunit | | Authors: | Twomey, E.C, Yelshanskaya, M.V, Vassilevski, A.A, Sobolevsky, A.I. | | Deposit date: | 2018-06-04 | | Release date: | 2018-08-22 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Mechanisms of Channel Block in Calcium-Permeable AMPA Receptors.
Neuron, 99, 2018
|
|
2MQU
 
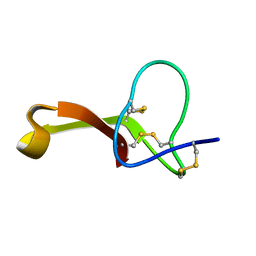 | | Spatial structure of Hm-3, a membrane-active spider toxin affecting sodium channels | | Descriptor: | Neurotoxin Hm-3 | | Authors: | Myshkin, M.Y, Shenkarev, Z.O, Paramonov, A.S, Berkut, A.A, Grishin, E.V, Vassilevski, A.A. | | Deposit date: | 2014-06-27 | | Release date: | 2014-11-05 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structure of membrane-active toxin from crab spider Heriaeus melloteei suggests parallel evolution of sodium channel gating modifiers in Araneomorphae and Mygalomorphae.
J. Biol. Chem., 290, 2015
|
|
2M6A
 
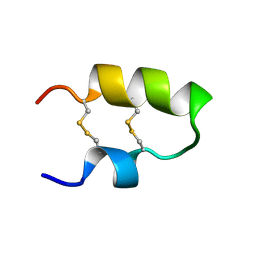 | | NMR spatial structure of the antimicrobial peptide Tk-Amp-X2 | | Descriptor: | Predicted protein | | Authors: | Usmanova, D.R, Mineev, K.S, Arseniev, A.S, Berkut, A.A, Oparin, P.B, Grishin, E.V, Vassilevski, A.A. | | Deposit date: | 2013-03-28 | | Release date: | 2014-04-02 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structural similarity between defense peptide from wheat and scorpion neurotoxin permits rational functional design
J.Biol.Chem., 289, 2014
|
|
2L2R
 
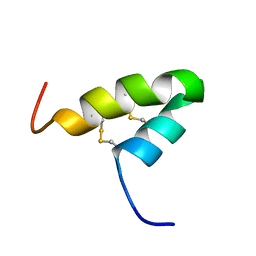 | | Helical hairpin structure of a novel antimicrobial peptide EcAMP1 from seeds of barnyard grass (Echinochloa crus-galli) | | Descriptor: | Antimicrobial peptide EcAMP1 | | Authors: | Nolde, S.B, Barinov, N.A, Balashova, T.A, Arseniev, A.S, Vassilevski, A.A, Rogozhin, E.A, Egorov, T.A, Grishin, E.V. | | Deposit date: | 2010-08-26 | | Release date: | 2011-05-11 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Disulfide-stabilized Helical Hairpin Structure and Activity of a Novel Antifungal Peptide EcAMP1 from Seeds of Barnyard Grass (Echinochloa crus-galli).
J.Biol.Chem., 286, 2011
|
|
2LQX
 
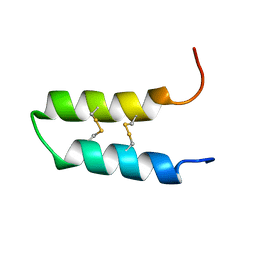 | | NMR spatial structure of the trypsin inhibitor BWI-2c from the buckwheat seeds | | Descriptor: | Trypsin inhibitor BWI-2c | | Authors: | Mineev, K.S, Vassilevski, A.A, Oparin, P.B, Grishin, E.V, Egorov, T.A. | | Deposit date: | 2012-03-19 | | Release date: | 2012-05-30 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Buckwheat trypsin inhibitor with helical hairpin structure belongs to a new family of plant defence peptides.
Biochem.J., 446, 2012
|
|
2N1S
 
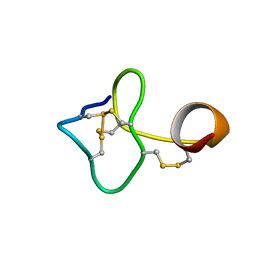 | |
7OXF
 
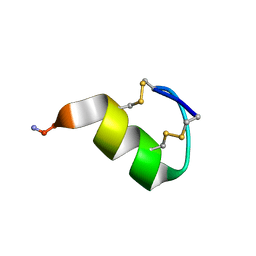 | |
2KGU
 
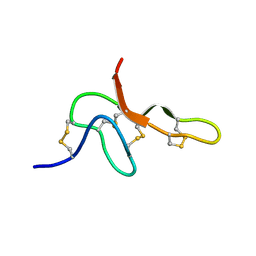 | | Spatial structure of purotoxin-1 in water | | Descriptor: | Purotoxin-1 | | Authors: | Nadezhdin, K, Vassilevski, A, Arseniev, A, Grishin, E, Pluzhnikov, K, Korolkova, Y. | | Deposit date: | 2009-03-20 | | Release date: | 2010-03-31 | | Last modified: | 2019-05-15 | | Method: | SOLUTION NMR | | Cite: | Novel peptide from spider venom inhibits P2X3 receptors and inflammatory pain.
Ann Neurol, 67, 2010
|
|
2MZF
 
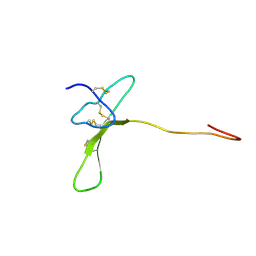 | | Purotoxin-2 NMR structure in water | | Descriptor: | Purotoxin-2 | | Authors: | Nadezhdin, K, Vassilevski, A, Oparin, P, Grishin, E, Arseniev, A. | | Deposit date: | 2015-02-12 | | Release date: | 2016-04-13 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure of purotoxin-2 from wolf spider: modular design and membrane-assisted mode of action in arachnid toxins.
Biochem. J., 473, 2016
|
|
2MZG
 
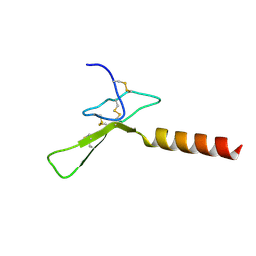 | | Purotoxin-2 NMR structure in DPC micelles | | Descriptor: | Purotoxin-2 | | Authors: | Nadezhdin, K, Oparin, P, Vassilevski, A, Grishin, E, Arseniev, A. | | Deposit date: | 2015-02-12 | | Release date: | 2016-04-13 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure of purotoxin-2 from wolf spider: modular design and membrane-assisted mode of action in arachnid toxins.
Biochem. J., 473, 2016
|
|
6F61
 
 | |
2N85
 
 | | NMR structure of OtTx1a - AMP in DPC micelles | | Descriptor: | Spiderine-1a | | Authors: | Nadezhdin, K, Romanovskaya, D, Sachkova, M, Vassilevski, A, Grishin, E, Kovalchuk, S, Arseniev, A. | | Deposit date: | 2015-10-05 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Modular toxin from the lynx spider Oxyopes takobius: Structure of spiderine domains in solution and membrane-mimicking environment.
Protein Sci., 26, 2017
|
|
2N86
 
 | | NMR structure of OtTx1a - ICK | | Descriptor: | Spiderine-1a | | Authors: | Nadezhdin, K, Romanovskaya, D, Sachkova, M, Vassilevski, A, Grishin, E, Kovalchuk, S, Arseniev, A. | | Deposit date: | 2015-10-05 | | Release date: | 2016-10-19 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Modular toxin from the lynx spider Oxyopes takobius: Structure of spiderine domains in solution and membrane-mimicking environment.
Protein Sci., 26, 2017
|
|
