2JQX
 
 | | Solution structure of Malate Synthase G from joint refinement against NMR and SAXS data | | Descriptor: | Malate synthase G | | Authors: | Grishaev, A, Tugarinov, V, Kay, L.E, Trewhella, J, Bax, A. | | Deposit date: | 2007-06-13 | | Release date: | 2007-07-10 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Refined solution structure of the 82-kDa enzyme malate synthase G from joint NMR and synchrotron SAXS restraints
J.Biomol.Nmr, 40, 2008
|
|
5K6P
 
 | | The NMR structure of the m domain tri-helix bundle and C2 of human cardiac Myosin Binding Protein C | | Descriptor: | Myosin-binding protein C, cardiac-type | | Authors: | Michie, K.A, Kwan, A.H, Tung, C.S, Guss, J.M, Trewhella, J. | | Deposit date: | 2016-05-25 | | Release date: | 2016-11-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A Highly Conserved Yet Flexible Linker Is Part of a Polymorphic Protein-Binding Domain in Myosin-Binding Protein C.
Structure, 24, 2016
|
|
3FYR
 
 | | Crystal structure of the sporulation histidine kinase inhibitor Sda from Bacillus subtilis | | Descriptor: | Sporulation inhibitor sda | | Authors: | Jacques, D.A, Streamer, M, King, G.F, Guss, J.M, Trewhella, J, Langley, D.B. | | Deposit date: | 2009-01-23 | | Release date: | 2009-06-23 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure of the sporulation histidine kinase inhibitor Sda from Bacillus subtilis and insights into its solution state
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2JPW
 
 | | Solution structure of the bisphosphorylated cardiac specific N-extension of cardiac troponin I | | Descriptor: | Troponin I, cardiac muscle | | Authors: | Howarth, J.W, Rosevear, P.R, Meller, J, Solaro, R.J, Trewhella, J. | | Deposit date: | 2007-05-24 | | Release date: | 2007-12-11 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Phosphorylation-dependent conformational transition of the cardiac specific N-extension of troponin I in cardiac troponin
J.Mol.Biol., 373, 2007
|
|
1WHF
 
 | | COAGULATION FACTOR, NMR, 15 STRUCTURES | | Descriptor: | COAGULATION FACTOR X | | Authors: | Sunnerhagen, M, Olah, G.A, Stenflo, J, Forsen, S, Drakenberg, T, Trewhella, J. | | Deposit date: | 1996-06-18 | | Release date: | 1997-05-15 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | The relative orientation of Gla and EGF domains in coagulation factor X is altered by Ca2+ binding to the first EGF domain. A combined NMR-small angle X-ray scattering study.
Biochemistry, 35, 1996
|
|
1WHE
 
 | | COAGULATION FACTOR, NMR, 20 STRUCTURES | | Descriptor: | COAGULATION FACTOR X | | Authors: | Sunnerhagen, M, Olah, G.A, Stenflo, J, Forsen, S, Drakenberg, T, Trewhella, J. | | Deposit date: | 1996-06-18 | | Release date: | 1997-05-15 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | The relative orientation of Gla and EGF domains in coagulation factor X is altered by Ca2+ binding to the first EGF domain. A combined NMR-small angle X-ray scattering study.
Biochemistry, 35, 1996
|
|
3KWX
 
 | | Chemically modified Taka alpha-amylase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-amylase A type-1/2, CALCIUM ION | | Authors: | Siddiqui, K.S, Harrop, S.J, Poljak, A, De Francisci, D, Guerriero, G, Pilak, O, Burg, D, Raftery, M.J, Parkin, D.M, Trewhella, J, Cavicchioli, R. | | Deposit date: | 2009-12-01 | | Release date: | 2009-12-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Modified alpha-amylase with a molten-globule state has enhanced thermal stability
To be Published
|
|
2A5M
 
 | |
2KWA
 
 | | 1H, 13C and 15N backbone and side chain resonance assignments of the N-terminal domain of the histidine kinase inhibitor KipI from Bacillus subtilis | | Descriptor: | Kinase A inhibitor | | Authors: | Hynson, R.M.G, Kwan, A, Jacques, D.A, Mackay, J.P, Trewhella, J. | | Deposit date: | 2010-03-31 | | Release date: | 2010-11-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A Novel Structure of an Antikinase and its Inhibitor
J.Mol.Biol., 2010
|
|
3OPF
 
 | | Crystal structure of TTHA0988 in space group P212121 | | Descriptor: | GLYCEROL, Putative uncharacterized protein TTHA0988, SULFATE ION | | Authors: | Jacques, D.A, Kuramitsu, S, Yokoyama, S, Trewhella, J, Guss, J.M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-09-01 | | Release date: | 2011-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The structure of TTHA0988 from Thermus thermophilus, a KipI-KipA homologue incorrectly annotated as an allophanate hydrolase
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3OEP
 
 | | Crystal structure of TTHA0988 in space group P43212 | | Descriptor: | GLYCEROL, Putative uncharacterized protein TTHA0988, SULFATE ION | | Authors: | Jacques, D.A, Langley, D.B, Trewhella, J, Guss, J.M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-08-13 | | Release date: | 2010-11-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The structure of TTHA0988 from Thermus thermophilus, a KipI-KipA homologue incorrectly annotated as an allophanate hydrolase
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3ORE
 
 | | Crystal structure of TTHA0988 in space group P6522 | | Descriptor: | Putative uncharacterized protein TTHA0988 | | Authors: | Jacques, D.A, Kuramitsu, S, Yokoyama, S, Trewhella, J, Guss, J.M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-09-07 | | Release date: | 2011-02-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure of TTHA0988 from Thermus thermophilus, a KipI-KipA homologue incorrectly annotated as an allophanate hydrolase
Acta Crystallogr.,Sect.D, 67, 2011
|
|
5HFS
 
 | | CRYSTAL STRUCTURE OF C-TERMINAL DOMAIN OF CARGO PROTEINS OF TYPE IX SECRETION SYSTEM | | Descriptor: | CALCIUM ION, Gingipain R2, ZINC ION | | Authors: | Golik, P, Szmigielski, B, Ksiazek, M, Nowakowska, Z, Mizgalska, D, Nowak, M, Dubin, G, Potempa, J. | | Deposit date: | 2016-01-07 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The outer-membrane export signal of Porphyromonas gingivalis type IX secretion system (T9SS) is a conserved C-terminal beta-sandwich domain.
Sci Rep, 6, 2016
|
|
6FGA
 
 | | Crystal structure of TRIM21 E3 ligase, RING domain in complex with its cognate E2 conjugating enzyme UBE2E1 | | Descriptor: | E3 ubiquitin-protein ligase TRIM21, GLYCEROL, Ubiquitin-conjugating enzyme E2 E1, ... | | Authors: | Anandapadamanaban, M, Moche, M, Sunnerhagen, M. | | Deposit date: | 2018-01-10 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | E3 ubiquitin-protein ligase TRIM21-mediated lysine capture by UBE2E1 reveals substrate-targeting mode of a ubiquitin-conjugating E2.
J.Biol.Chem., 294, 2019
|
|
2ZP2
 
 | |
5AG8
 
 | | CRYSTAL STRUCTURE OF A MUTANT (665I6H) OF THE C-TERMINAL DOMAIN OF RGPB | | Descriptor: | GINGIPAIN R2, GLYCEROL, SULFATE ION | | Authors: | de Diego, I, Ksiazek, M, Mizgalska, D, Golik, P, Szmigielski, B, Nowak, M, Nowakowska, Z, Potempa, B, Koneru, L, Nguyen, K.A, Enghild, J, Thogersen, I.B, Dubin, G, Gomis-Ruth, F.X, Potempa, J. | | Deposit date: | 2015-01-29 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Outer-Membrane Export Signal of Porphyromonas Gingivalis Type Ix Secretion System (T9Ss) is a Conserved C-Terminal Beta-Sandwich Domain.
Sci.Rep., 6, 2016
|
|
5AG9
 
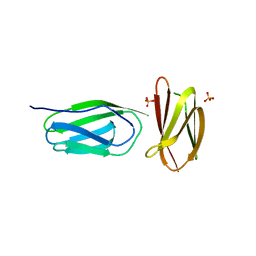 | | CRYSTAL STRUCTURE OF A MUTANT (665sXa) C-TERMINAL DOMAIN OF RGPB | | Descriptor: | Gingipain R2, SULFATE ION | | Authors: | de Diego, I, Ksiazek, M, Mizgalska, D, Golik, P, Szmigielski, B, Nowak, M, Nowakowska, Z, Potempa, B, Koneru, L, Nguyen, K.A, Enghild, J, Thogersen, I.B, Dubin, G, Gomis-Ruth, F.X, Potempa, J. | | Deposit date: | 2015-01-29 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | The outer-membrane export signal of Porphyromonas gingivalis type IX secretion system (T9SS) is a conserved C-terminal beta-sandwich domain.
Sci Rep, 6, 2016
|
|
4WIJ
 
 | | HUMAN SPLICING FACTOR, CONSTRUCT 1 | | Descriptor: | Splicing factor, proline- and glutamine-rich | | Authors: | lee, M, bond, c.s. | | Deposit date: | 2014-09-26 | | Release date: | 2015-04-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | The structure of human SFPQ reveals a coiled-coil mediated polymer essential for functional aggregation in gene regulation.
Nucleic Acids Res., 43, 2015
|
|
4WII
 
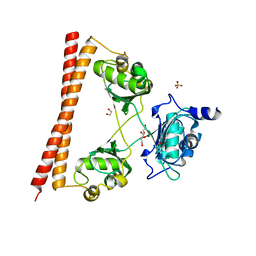 | | HUMAN SPLICING FACTOR, CONSTRUCT 3 | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Splicing factor, ... | | Authors: | lee, M, bond, c.s. | | Deposit date: | 2014-09-26 | | Release date: | 2015-04-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The structure of human SFPQ reveals a coiled-coil mediated polymer essential for functional aggregation in gene regulation.
Nucleic Acids Res., 43, 2015
|
|
4WIK
 
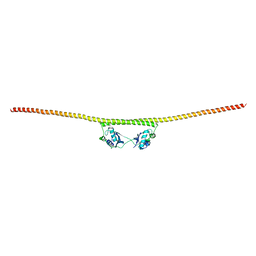 | | HUMAN SPLICING FACTOR, CONSTRUCT 2 | | Descriptor: | Splicing factor, proline- and glutamine-rich | | Authors: | lee, M, bond, c.s. | | Deposit date: | 2014-09-26 | | Release date: | 2015-04-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of human SFPQ reveals a coiled-coil mediated polymer essential for functional aggregation in gene regulation.
Nucleic Acids Res., 43, 2015
|
|
6CRL
 
 | | HusA haemophore from Porphyromonas gingivalis | | Descriptor: | hypothetical protein PG_2227 | | Authors: | Gell, D.A, Kwan, A.H, Horne, J, Hugrass, B.M, Collins, D.A.T. | | Deposit date: | 2018-03-19 | | Release date: | 2018-10-10 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural properties of a haemophore facilitate targeted elimination of the pathogen Porphyromonas gingivalis.
Nat Commun, 9, 2018
|
|
6BQS
 
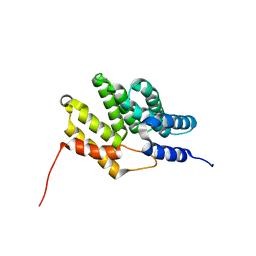 | | HusA haemophore from Porphyromonas gingivalis | | Descriptor: | hypothetical protein PG_2227 | | Authors: | Gell, D.A, Kwan, A.H, Horne, J, Hugrass, B.M, Collins, D.A.T. | | Deposit date: | 2017-11-28 | | Release date: | 2018-10-10 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural properties of a haemophore facilitate targeted elimination of the pathogen Porphyromonas gingivalis.
Nat Commun, 9, 2018
|
|
3MMK
 
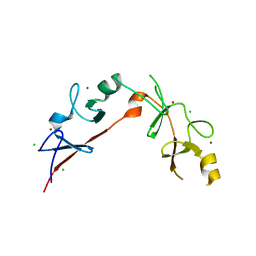 | | The structural basis for partial redundancy in a class of transcription factors, the lim-homeodomain proteins, in neural cell type specification | | Descriptor: | CHLORIDE ION, Fusion of LIM/homeobox protein Lhx4, linker, ... | | Authors: | Gadd, M.S, Langley, D.B, Guss, J.M, Matthews, J.M. | | Deposit date: | 2010-04-20 | | Release date: | 2011-07-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.157 Å) | | Cite: | The structural basis for partial redundancy in a class of transcription factors, the lim-homeodomain proteins, in neural cell type specification.
J.Biol.Chem., 2011
|
|
4ZZL
 
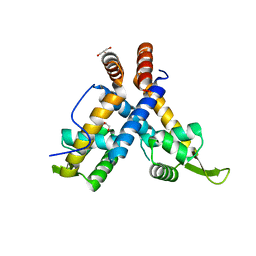 | | MexR R21W derepressor mutant causing multidrug resistance in P. aeruginosa by mexAB-oprM efflux pump expression | | Descriptor: | GLYCEROL, MULTIDRUG RESISTANCE OPERON REPRESSOR | | Authors: | Anandapadamanaban, M, Pilstal, R, Ziauddin, J.M.E, Moche, M, Wallner, B, Sunnerhagen, M. | | Deposit date: | 2015-04-10 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Mutation-Induced Population Shift in the Mexr Conformational Ensemble Disengages DNA Binding: A Novel Mechanism for Marr Family Derepression.
Structure, 24, 2016
|
|
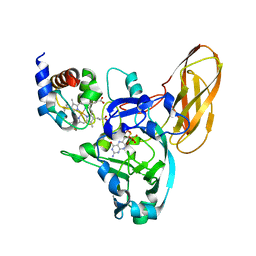
4PW9
 
 | |
