1W3K
 
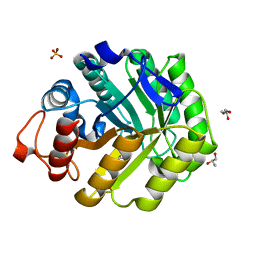 | | ENDOGLUCANASE CEL5A FROM BACILLUS AGARADHAERENS IN COMPLEX WITH CELLOBIO DERIVED-TETRAHYDROOXAZINE | | Descriptor: | ENDOGLUCANASE 5A, GLYCEROL, SULFATE ION, ... | | Authors: | Gloster, T.M, Macdonald, J.M, Tarling, C.A, Stick, R.V, Withers, S.W, Davies, G.J. | | Deposit date: | 2004-07-16 | | Release date: | 2004-09-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural, Thermodynamic, and Kinetic Analyses of Tetrahydrooxazine-Derived Inhibitors Bound to {Beta}-Glucosidases
J.Biol.Chem., 279, 2004
|
|
1W3L
 
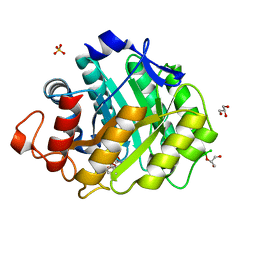 | | ENDOGLUCANASE CEL5A FROM BACILLUS AGARADHAERENS IN COMPLEX WITH CELLOTRI DERIVED-TETRAHYDROOXAZINE | | Descriptor: | ENDOGLUCANASE 5A, GLYCEROL, SULFATE ION, ... | | Authors: | Gloster, T.M, Macdonald, J.M, Tarling, C.A, Stick, R.V, Withers, S.W, Davies, G.J. | | Deposit date: | 2004-07-16 | | Release date: | 2004-09-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Structural, Thermodynamic, and Kinetic Analyses of Tetrahydrooxazine-Derived Inhibitors Bound to {Beta}-Glucosidases
J.Biol.Chem., 279, 2004
|
|
1W3J
 
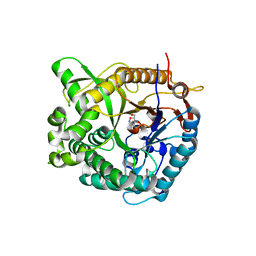 | | Family 1 b-glucosidase from Thermotoga maritima in complex with tetrahydrooxazine | | Descriptor: | BETA-GLUCOSIDASE, TETRAHYDROOXAZINE | | Authors: | Gloster, T.M, Macdonald, J.M, Tarling, C.A, Stick, R.V, Withers, S.W, Davies, G.J. | | Deposit date: | 2004-07-16 | | Release date: | 2004-09-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural, Thermodynamic, and Kinetic Analyses of Tetrahydrooxazine-Derived Inhibitors Bound to {Beta}-Glucosidases
J.Biol.Chem., 279, 2004
|
|
1V0K
 
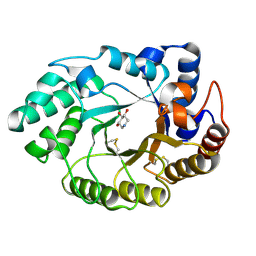 | | Xylanase Xyn10A from Streptomyces lividans in complex with xylobio-deoxynojirimycin at pH 5.8 | | Descriptor: | ENDO-1,4-BETA-XYLANASE A, PIPERIDINE-3,4,5-TRIOL, beta-D-xylopyranose | | Authors: | Gloster, T.M, Williams, S.J, Roberts, S, Tarling, C.A, Wicki S, J, Withers, G, Davies, G.J. | | Deposit date: | 2004-03-31 | | Release date: | 2004-08-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Atomic Resolution Analyses of the Binding of Xylobiose-Derived Deoxynojirimycin and Isofagomine to Xylanase Xyn10A
Chem.Commun.(Camb.), 16, 2004
|
|
1V0M
 
 | | Xylanase Xyn10a from Streptomyces lividans in complex with xylobio-deoxynojirimycin at pH 7.5 | | Descriptor: | ENDO-1,4-BETA-XYLANASE A, IMIDAZOLE, PIPERIDINE-3,4,5-TRIOL, ... | | Authors: | Gloster, T.M, Williams, S.J, Roberts, S, Tarling, C.A, Wicki, J, Withers, S.G, Davies, G.J. | | Deposit date: | 2004-03-31 | | Release date: | 2004-08-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Atomic Resolution Analyses of the Binding of Xylobiose-Derived Deoxynojirimycin and Isofagomine to Xylanase Xyn10A
Chem.Commun.(Camb.), 16, 2004
|
|
1V0L
 
 | | Xylanase Xyn10A from Streptomyces lividans in complex with xylobio-isofagomine at pH 5.8 | | Descriptor: | ENDO-1,4-BETA-XYLANASE A, PIPERIDINE-3,4-DIOL, beta-D-xylopyranose | | Authors: | Gloster, T.M, Williams, S.J, Roberts, S, Tarling, C.A, Wicki, J, Withers, S.G, Davies, G.J. | | Deposit date: | 2004-03-31 | | Release date: | 2004-08-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Atomic Resolution Analyses of the Binding of Xylobiose-Derived Deoxynojirimycin and Isofagomine to Xylanase Xyn10A
Chem.Commun.(Camb.), 16, 2004
|
|
1V0N
 
 | | Xylanase Xyn10a from Streptomyces lividans in complex with xylobio-isofagomine at pH 7.5 | | Descriptor: | 1,2-ETHANEDIOL, ENDO-1,4-BETA-XYLANASE A, IMIDAZOLE, ... | | Authors: | Gloster, T.M, Williams, S.J, Roberts, S, Tarling, C.A, Wicki, J, Withers, S.G, Davies, G.J. | | Deposit date: | 2004-03-31 | | Release date: | 2004-08-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic Resolution Analyses of the Binding of Xylobiose-Derived Deoxynojirimycin and Isofagomine to Xylanase Xyn10A
Chem.Commun.(Camb.), 16, 2004
|
|
1UQU
 
 | | Trehalose-6-phosphate from E. coli bound with UDP-glucose. | | Descriptor: | ALPHA, ALPHA-TREHALOSE-PHOSPHATE SYNTHASE, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Gibson, R.P, Tarling, C.A, Roberts, S, Withers, S.G, Davies, G.J. | | Deposit date: | 2003-10-20 | | Release date: | 2003-11-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The donor subsite of trehalose-6-phosphate synthase: binary complexes with UDP-glucose and UDP-2-deoxy-2-fluoro-glucose at 2 A resolution.
J. Biol. Chem., 279, 2004
|
|
1UQT
 
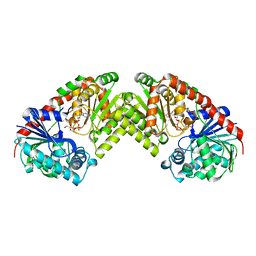 | | Trehalose-6-phosphate from E. coli bound with UDP-2-fluoro glucose. | | Descriptor: | ALPHA, ALPHA-TREHALOSE-PHOSPHATE SYNTHASE, URIDINE-5'-DIPHOSPHATE-2-DEOXY-2-FLUORO-ALPHA-D-GLUCOSE | | Authors: | Gibson, R.P, Tarling, C.A, Roberts, S, Withers, S.G, Davies, G.J. | | Deposit date: | 2003-10-20 | | Release date: | 2003-12-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The donor subsite of trehalose-6-phosphate synthase: binary complexes with UDP-glucose and UDP-2-deoxy-2-fluoro-glucose at 2 A resolution.
J. Biol. Chem., 279, 2004
|
|
2C1X
 
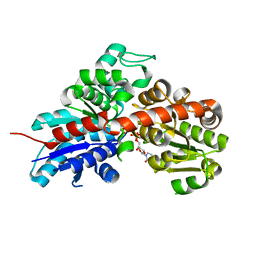 | | Structure and activity of a flavonoid 3-O glucosyltransferase reveals the basis for plant natural product modification | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, UDP-GLUCOSE FLAVONOID 3-O GLYCOSYLTRANSFERASE, URIDINE-5'-DIPHOSPHATE | | Authors: | Offen, W, Martinez-Fleites, C, Kiat-Lim, E, Yang, M, Davis, B.G, Tarling, C.A, Ford, C.M, Bowles, D.J, Davies, G.J. | | Deposit date: | 2005-09-22 | | Release date: | 2006-01-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a Flavonoid Glucosyltransferase Reveals the Basis for Plant Natural Product Modification.
Embo J., 25, 2006
|
|
2C1Z
 
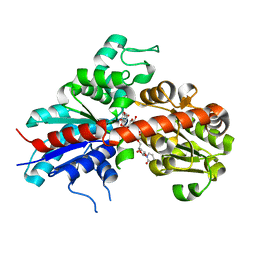 | | Structure and activity of a flavonoid 3-O glucosyltransferase reveals the basis for plant natural product modification | | Descriptor: | 3,5,7-TRIHYDROXY-2-(4-HYDROXYPHENYL)-4H-CHROMEN-4-ONE, UDP-GLUCOSE FLAVONOID 3-O GLYCOSYLTRANSFERASE, URIDINE-5'-DIPHOSPHATE-2-DEOXY-2-FLUORO-ALPHA-D-GLUCOSE | | Authors: | Offen, W, Martinez-Fleites, C, Kiat-Lim, E, Yang, M, Davis, B.G, Tarling, C.A, Ford, C.M, Bowles, D.J, Davies, G.J. | | Deposit date: | 2005-09-22 | | Release date: | 2006-01-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a Flavonoid Glucosyltransferase Reveals the Basis for Plant Natural Product Modification.
Embo J., 25, 2006
|
|
2C9Z
 
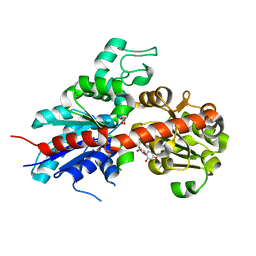 | | Structure and activity of a flavonoid 3-0 glucosyltransferase reveals the basis for plant natural product modification | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, UDP GLUCOSE:FLAVONOID 3-O-GLUCOSYLTRANSFERASE, URIDINE-5'-DIPHOSPHATE | | Authors: | Offen, W, Martinez-Fleites, C, Kiat-Lim, E, Yang, M, Davis, B.G, Tarling, C.A, Ford, C.M, Bowles, D.J, Davies, G.J. | | Deposit date: | 2005-12-15 | | Release date: | 2006-01-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a Flavonoid Glucosyltransferase Reveals the Basis for Plant Natural Product Modification.
Embo J., 25, 2006
|
|
2V38
 
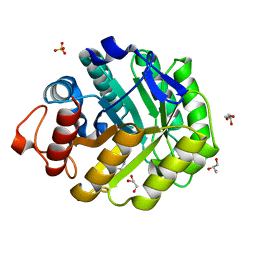 | | Family 5 endoglucanase Cel5A from Bacillus agaradhaerens in complex with cellobio-derived noeuromycin | | Descriptor: | (3S,4R,5R)-3,4-DIHYDROXY-5-(HYDROXYMETHYL)PIPERIDIN-2-ONE, ENDOGLUCANASE 5A, GLYCEROL, ... | | Authors: | Gloster, T.M, Meloncelli, P.J, Money, V.A, Tarling, C.A, Davies, G.J, Withers, S.G, Stick, R.V. | | Deposit date: | 2007-06-13 | | Release date: | 2007-06-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | D-Glucosylated Derivatives of Isofagomine and Noeuromycin and Their Potential as Inhibitors of Beta-Glycoside Hydrolases
To be Published
|
|
2V3G
 
 | | Structure of a family 26 lichenase in complex with noeuromycin | | Descriptor: | (2R,3S,4R,5R)-5-(HYDROXYMETHYL)PIPERIDINE-2,3,4-TRIOL, ENDOGLUCANASE H, beta-D-glucopyranose | | Authors: | Meloncelli, P.J, Gloster, T.M, Money, V.A, Tarling, C.A, Davies, G.J, Withers, S.G, Stick, R.V. | | Deposit date: | 2007-06-18 | | Release date: | 2007-09-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | D-Glucosylated Derivatives of Isofagomine and Noeuromycin and Their Potential as Inhibitors of Beta-Glycoside Hydrolases
To be Published
|
|
2WLZ
 
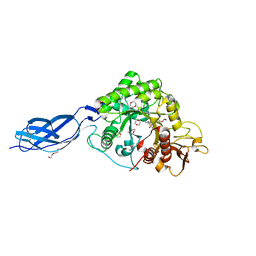 | | Chitinase A from Serratia marcescens ATCC990 in complex with Chitobio- thiazoline. | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, ... | | Authors: | Taylor, E.J, Dennis, R.J, Macdonald, J.M, Tarling, C.A, Knapp, S, Withers, S.G, Davies, G.J. | | Deposit date: | 2009-06-29 | | Release date: | 2010-03-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Chitinase Inhibition by Chitobiose and Chitotriose Thiazolines.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
2WLY
 
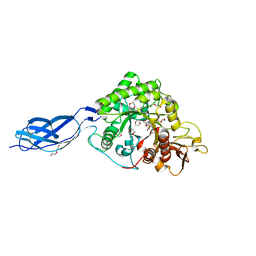 | | Chitinase A from Serratia marcescens ATCC990 in complex with Chitotrio-thiazoline. | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 2-deoxy-2-(ethanethioylamino)-beta-D-glucopyranose, 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, ... | | Authors: | Taylor, E.J, Dennis, R.J, Macdonald, J.M, Tarling, C.A, Knapp, S, Withers, S.G, Davies, G.J. | | Deposit date: | 2009-06-29 | | Release date: | 2010-03-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Chitinase inhibition by chitobiose and chitotriose thiazolines.
Angew. Chem. Int. Ed. Engl., 49, 2010
|
|
2WM0
 
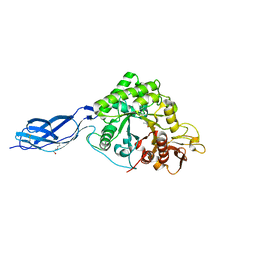 | | Chitinase A from Serratia marcescens ATCC990 in complex with Chitobio- thiazoline thioamide. | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, ... | | Authors: | Taylor, E.J, Dennis, R.J, Macdonald, J.M, Tarling, C.A, Knapp, S, Withers, S.G, Davies, G.J. | | Deposit date: | 2009-06-29 | | Release date: | 2010-03-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Chitinase Inhibition by Chitobiose and Chitotriose Thiazolines.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
2WK2
 
 | | Chitinase A from Serratia marcescens ATCC990 in complex with Chitotrio-thiazoline dithioamide. | | Descriptor: | 2-deoxy-2-(ethanethioylamino)-beta-D-glucopyranose-(1-4)-2-deoxy-2-(ethanethioylamino)-beta-D-glucopyranose, 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, CHITINASE A, ... | | Authors: | Taylor, E.J, Dennis, R.J, Macdonald, J.M, Tarling, C.A, Knapp, S, Withers, S.G, Davies, G.J. | | Deposit date: | 2009-06-04 | | Release date: | 2010-03-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Chitinase Inhibition by Chitobiose and Chitotriose Thiazolines.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
1OJJ
 
 | | Anatomy of glycosynthesis: Structure and kinetics of the Humicola insolens Cel7BE197A and E197S glycosynthase mutants | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDOGLUCANASE I, beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Ducros, V.M.-A, Tarling, C.A, Zechel, D.L, Brzozowski, A.M, Frandsen, T.P, Von Ossowski, I, Schulein, M, Withers, S.G, Davies, G.J. | | Deposit date: | 2003-07-10 | | Release date: | 2004-01-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Anatomy of Glycosynthesis: Structure and Kinetics of the Humicola Insolens Cel7B E197A and E197S Glycosynthase Mutants
Chem.Biol., 10, 2003
|
|
1OJK
 
 | | Anatomy of glycosynthesis: Structure and kinetics of the Humicola insolens Cel7BE197A and E197S glycosynthase mutants | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDOGLUCANASE I, GLYCEROL, ... | | Authors: | Ducros, V.M.-A, Tarling, C.A, Zechel, D.L, Brzozowski, A.M, Frandsen, T.P, Von Ossowski, I, Schulein, M, Withers, S.G, Davies, G.J. | | Deposit date: | 2003-07-10 | | Release date: | 2004-01-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Anatomy of Glycosynthesis: Structure and Kinetics of the Humicola Insolens Cel7B E197A and E197S Glycosynthase Mutants
Chem.Biol., 10, 2003
|
|
1OJI
 
 | | Anatomy of glycosynthesis: Structure and kinetics of the Humicola insolens Cel7B E197A and E197S glycosynthase mutants | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDOGLUCANASE I, GLYCEROL | | Authors: | Ducros, V.M.-A, Tarling, C.A, Zechel, D.L, Brzozowski, A.M, Frandsen, T.P, Von Ossowski, I, Schulein, M, Withers, S.G, Davies, G.J. | | Deposit date: | 2003-07-10 | | Release date: | 2004-01-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Anatomy of Glycosynthesis: Structure and Kinetics of the Humicola Insolens Cel7B E197A and E197S Glycosynthase Mutants
Chem.Biol., 10, 2003
|
|
3D52
 
 | | GOLGI MANNOSIDASE II complex with an N-aryl carbamate derivative of gluco-hydroxyiminolactam | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Alpha-mannosidase 2, ZINC ION, ... | | Authors: | Kuntz, D.A, Tarling, C.A, Withers, S.G, Rose, D.R. | | Deposit date: | 2008-05-15 | | Release date: | 2008-08-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural analysis of Golgi alpha-mannosidase II inhibitors identified from a focused glycosidase inhibitor screen.
Biochemistry, 47, 2008
|
|
3D50
 
 | | GOLGI MANNOSIDASE II complex with N-octyl-6-epi-valienamine | | Descriptor: | (1S,2S,3R,6R)-4-(hydroxymethyl)-6-(octylamino)cyclohex-4-ene-1,2,3-triol, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kuntz, D.A, Tarling, C.A, Withers, S.G, Rose, D.R. | | Deposit date: | 2008-05-15 | | Release date: | 2008-08-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural analysis of Golgi alpha-mannosidase II inhibitors identified from a focused glycosidase inhibitor screen.
Biochemistry, 47, 2008
|
|
3D4Y
 
 | | GOLGI MANNOSIDASE II complex with mannoimidazole | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, (5R,6R,7S,8R)-5-(HYDROXYMETHYL)-5,6,7,8-TETRAHYDROIMIDAZO[1,2-A]PYRIDINE-6,7,8-TRIOL, ... | | Authors: | Kuntz, D.A, Tarling, C.A, Withers, S.G, Rose, D.R. | | Deposit date: | 2008-05-15 | | Release date: | 2008-08-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structural analysis of Golgi alpha-mannosidase II inhibitors identified from a focused glycosidase inhibitor screen.
Biochemistry, 47, 2008
|
|
3D4Z
 
 | | GOLGI MANNOSIDASE II complex with gluco-imidazole | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Alpha-mannosidase 2, GLUCOIMIDAZOLE, ... | | Authors: | Kuntz, D.A, Tarling, C.A, Withers, S.G, Rose, D.R. | | Deposit date: | 2008-05-15 | | Release date: | 2008-08-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structural analysis of Golgi alpha-mannosidase II inhibitors identified from a focused glycosidase inhibitor screen.
Biochemistry, 47, 2008
|
|
