2D73
 
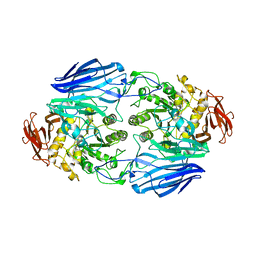 | | Crystal Structure Analysis of SusB | | Descriptor: | CALCIUM ION, alpha-glucosidase SusB | | Authors: | Kitamura, M, Yao, M. | | Deposit date: | 2005-11-15 | | Release date: | 2007-02-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and functional analysis of a glycoside hydrolase family 97 enzyme from Bacteroides thetaiotaomicron.
J.Biol.Chem., 283, 2008
|
|
7WKF
 
 | | Antimicrobial peptide-LaIT2 | | Descriptor: | Beta-KTx-like peptide LaIT2 | | Authors: | Tamura, M, Morita, H, Ohki, S. | | Deposit date: | 2022-01-09 | | Release date: | 2023-04-05 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Structural and functional studies of LaIT2, an antimicrobial and insecticidal peptide from Liocheles australasiae.
Toxicon, 214, 2022
|
|
3VOF
 
 | | Cellobiohydrolase mutant, CcCel6C D102A, in the closed form | | Descriptor: | Cellobiohydrolase, beta-D-glucopyranose | | Authors: | Tamura, M, Miyazaki, T, Tanaka, Y, Yoshida, M, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2012-01-23 | | Release date: | 2012-03-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Comparison of the structural changes in two cellobiohydrolases, CcCel6A and CcCel6C, from Coprinopsis cinerea - a tweezer-like motion in the structure of CcCel6C
Febs J., 279, 2012
|
|
3VOH
 
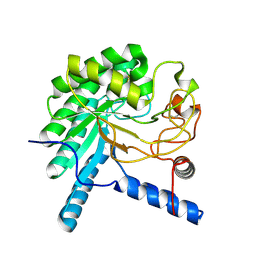 | | CcCel6A catalytic domain complexed with cellobiose | | Descriptor: | Cellobiohydrolase, beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Tamura, M, Miyazaki, T, Tanaka, Y, Yoshida, M, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2012-01-24 | | Release date: | 2012-03-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Comparison of the structural changes in two cellobiohydrolases, CcCel6A and CcCel6C, from Coprinopsis cinerea - a tweezer-like motion in the structure of CcCel6C
Febs J., 279, 2012
|
|
3VOJ
 
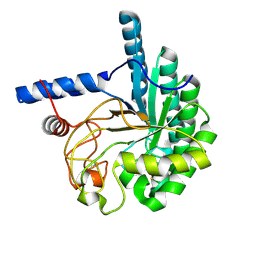 | | CcCel6A catalytic domain mutant D164A | | Descriptor: | Cellobiohydrolase | | Authors: | Tamura, M, Miyazaki, T, Tanaka, Y, Yoshida, M, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2012-01-24 | | Release date: | 2012-03-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Comparison of the structural changes in two cellobiohydrolases, CcCel6A and CcCel6C, from Coprinopsis cinerea - a tweezer-like motion in the structure of CcCel6C
Febs J., 279, 2012
|
|
3VOG
 
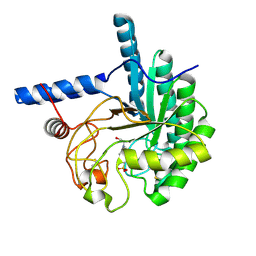 | | Catalytic domain of the cellobiohydrolase, CcCel6A, from Coprinopsis cinerea | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Cellobiohydrolase | | Authors: | Tamura, M, Miyazaki, T, Tanaka, Y, Yoshida, M, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2012-01-24 | | Release date: | 2012-03-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Comparison of the structural changes in two cellobiohydrolases, CcCel6A and CcCel6C, from Coprinopsis cinerea - a tweezer-like motion in the structure of CcCel6C
Febs J., 279, 2012
|
|
3VOI
 
 | | CcCel6A catalytic domain complexed with p-nitrophenyl beta-D-cellotrioside | | Descriptor: | 4-nitrophenyl beta-D-glucopyranosyl-(1->4)-beta-D-glucopyranosyl-(1->4)-beta-D-glucopyranoside, Cellobiohydrolase, MAGNESIUM ION | | Authors: | Tamura, M, Miyazaki, T, Tanaka, Y, Yoshida, M, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2012-01-24 | | Release date: | 2012-03-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Comparison of the structural changes in two cellobiohydrolases, CcCel6A and CcCel6C, from Coprinopsis cinerea - a tweezer-like motion in the structure of CcCel6C
Febs J., 279, 2012
|
|
2ZQ0
 
 | | Crystal structure of SusB complexed with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Alpha-glucosidase (Alpha-glucosidase SusB), CALCIUM ION | | Authors: | Yao, M, Tanaka, I, Kitamura, M. | | Deposit date: | 2008-07-31 | | Release date: | 2008-10-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and functional analysis of a glycoside hydrolase family 97 enzyme from Bacteroides thetaiotaomicron.
J.Biol.Chem., 283, 2008
|
|
4QCI
 
 | | PDGF-B blocking antibody bound to PDGF-BB | | Descriptor: | Platelet-derived growth factor subunit B, anti-PDGF-BB antibody - Light Chain, anti-PDGF-BB antibody - Heavy chain | | Authors: | Kuai, J, Mosyak, L, Tam, M, LaVallie, E, Pullen, N, Carven, G. | | Deposit date: | 2014-05-12 | | Release date: | 2015-03-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization of Binding Mode of Action of a Blocking Anti-Platelet-Derived Growth Factor (PDGF)-B Monoclonal Antibody, MOR8457, Reveals Conformational Flexibility and Avidity Needed for PDGF-BB To Bind PDGF Receptor-beta.
Biochemistry, 54, 2015
|
|
1R5Z
 
 | | Crystal Structure of Subunit C of V-ATPase | | Descriptor: | V-type ATP synthase subunit C | | Authors: | Iwata, M, Imamura, H, Stambouli, E, Ikeda, C, Tamakoshi, M, Nagata, K, Makyio, H, Hankamer, B, Barber, J, Yoshida, M, Yokoyama, K, Iwata, S. | | Deposit date: | 2003-10-14 | | Release date: | 2004-01-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of a central stalk subunit C and reversible association/dissociation of vacuole-type ATPase.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
4X9U
 
 | | Crystal structure of the kiwifruit allergen Act d 5 | | Descriptor: | Kiwellin | | Authors: | Offermann, L.R, Perdue, M.L, Giangrieco, I, Tamburrini, M, Ciardiello, M.A, Chruszcz, M. | | Deposit date: | 2014-12-11 | | Release date: | 2015-07-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Elusive Structural, Functional, and Immunological Features of Act d 5, the Green Kiwifruit Kiwellin.
J.Agric.Food Chem., 63, 2015
|
|
1UUH
 
 | | Hyaluronan binding domain of human CD44 | | Descriptor: | CD44 ANTIGEN | | Authors: | Teriete, P, Banerji, S, Noble, M, Blundell, C, Wright, A, Pickford, A, Lowe, E, Mahoney, D, Tammi, M, Kahmann, J, Campbell, I, Day, A, Jackson, D. | | Deposit date: | 2003-12-19 | | Release date: | 2004-03-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the Regulatory Hyaluronan-Binding Domain in the Inflammatory Leukocyte Homing Receptor Cd44
Mol.Cell, 13, 2004
|
|
1WE5
 
 | | Crystal Structure of Alpha-Xylosidase from Escherichia coli | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Putative family 31 glucosidase yicI | | Authors: | Ose, T, Kitamura, M, Okuyama, M, Mori, H, Kimura, A, Watanabe, N, Yao, M, Tanaka, I. | | Deposit date: | 2004-05-24 | | Release date: | 2005-02-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Alpha-Xylosidase from Escherichia coli
TO BE PUBLISHED
|
|
6ZJN
 
 | | Respiratory complex I from Thermus thermophilus, NADH dataset, minor state | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, IRON/SULFUR CLUSTER, NADH-quinone oxidoreductase subunit 1, ... | | Authors: | Kaszuba, K, Tambalo, M, Gallagher, G.T, Sazanov, L.A. | | Deposit date: | 2020-06-29 | | Release date: | 2020-09-02 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Key role of quinone in the mechanism of respiratory complex I.
Nat Commun, 11, 2020
|
|
6ZJL
 
 | | Respiratory complex I from Thermus thermophilus, NAD+ dataset, major state | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Kaszuba, K, Tambalo, M, Gallagher, G.T, Sazanov, L.A. | | Deposit date: | 2020-06-29 | | Release date: | 2020-09-02 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Key role of quinone in the mechanism of respiratory complex I.
Nat Commun, 11, 2020
|
|
2VPY
 
 | | Polysulfide reductase with bound quinone inhibitor, pentachlorophenol (PCP) | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, HYPOTHETICAL MEMBRANE SPANNING PROTEIN, IRON/SULFUR CLUSTER, ... | | Authors: | Jormakka, M, Yokoyama, K, Yano, T, Tamakoshi, M, Akimoto, S, Shimamura, T, Curmi, P, Iwata, S. | | Deposit date: | 2008-03-09 | | Release date: | 2008-06-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular Mechanism of Energy Conservation in Polysulfide Respiration.
Nat.Struct.Mol.Biol., 15, 2008
|
|
2VPW
 
 | | Polysulfide reductase with bound menaquinone | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, HYPOTHETICAL MEMBRANE SPANNING PROTEIN, IRON/SULFUR CLUSTER, ... | | Authors: | Jormakka, M, Yokoyama, K, Yano, T, Tamakoshi, M, Akimoto, S, Shimamura, T, Curmi, P, Iwata, S. | | Deposit date: | 2008-03-09 | | Release date: | 2008-06-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Molecular Mechanism of Energy Conservation in Polysulfide Respiration.
Nat.Struct.Mol.Biol., 15, 2008
|
|
6ZIY
 
 | | Respiratory complex I from Thermus thermophilus, NADH dataset, major state | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Kaszuba, K, Tambalo, M, Gallagher, G.T, Sazanov, L.A. | | Deposit date: | 2020-06-26 | | Release date: | 2020-09-02 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.25 Å) | | Cite: | Key role of quinone in the mechanism of respiratory complex I.
Nat Commun, 11, 2020
|
|
6ZJY
 
 | | Respiratory complex I from Thermus thermophilus, NAD+ dataset, minor state | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, IRON/SULFUR CLUSTER, NADH-quinone oxidoreductase subunit 1, ... | | Authors: | Kaszuba, K, Tambalo, M, Gallagher, G.T, Sazanov, L.A. | | Deposit date: | 2020-06-29 | | Release date: | 2020-09-02 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Key role of quinone in the mechanism of respiratory complex I.
Nat Commun, 11, 2020
|
|
2VPZ
 
 | | Polysulfide reductase native structure | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, HYPOTHETICAL MEMBRANE SPANNING PROTEIN, IRON/SULFUR CLUSTER, ... | | Authors: | Jormakka, M, Yokoyama, K, Yano, T, Tamakoshi, M, Akimoto, S, Shimamura, T, Curmi, P, Iwata, S. | | Deposit date: | 2008-03-09 | | Release date: | 2008-06-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular Mechanism of Energy Conservation in Polysulfide Respiration
Nat.Struct.Mol.Biol., 15, 2008
|
|
2VPX
 
 | | Polysulfide reductase with bound quinone (UQ1) | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, HYPOTHETICAL MEMBRANE SPANNING PROTEIN, IRON/SULFUR CLUSTER, ... | | Authors: | Jormakka, M, Yokoyama, K, Yano, T, Tamakoshi, M, Akimoto, S, Shimamura, T, Curmi, P, Iwata, S. | | Deposit date: | 2008-03-09 | | Release date: | 2008-06-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Molecular Mechanism of Energy Conservation in Polysulfide Respiration.
Nat.Struct.Mol.Biol., 15, 2008
|
|
1WST
 
 | | Crystal structure of multiple substrate aminotransferase (MsAT) from Thermococcus profundus | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, multiple substrate aminotransferase | | Authors: | Lee, W.C, Manabe, F, Nemoto, N, Tamakoshi, M, Tanokura, M, Yamagishi, A. | | Deposit date: | 2004-11-10 | | Release date: | 2005-10-25 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of multiple substrate aminotransferase (MsAT) from Thermococcus profundus
To be Published
|
|
2RLU
 
 | | The Three Dimensional Structure of the Moorella thermoacetica Selenocysteine Insertion Sequence RNA Hairpin and its Interaction with the Elongation factor SelB | | Descriptor: | RNA (5'-R(*GP*GP*UP*UP*GP*CP*GP*GP*GP*UP*CP*UP*CP*GP*CP*AP*AP*CP*C)-3') | | Authors: | Beribisky, A.V, Tavares, T.J, Amborski, A.N, Motamed, M, Johnson, A.E, Mark, T.L, Johnson, P.E. | | Deposit date: | 2007-08-21 | | Release date: | 2008-02-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional structure of the Moorella thermoacetica selenocysteine insertion sequence RNA hairpin and its interaction with the elongation factor SelB
Rna, 13, 2007
|
|
4P6Q
 
 | | The crystal structure of the Split End protein SHARP adds a new layer of complexity to proteins containing RNA Recognition Motifs | | Descriptor: | Msx2-interacting protein, SULFATE ION | | Authors: | Arieti, F, Gabus, C, Tambalo, M, Huet, T, Round, A, Thore, S. | | Deposit date: | 2014-03-25 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of the Split End protein SHARP adds a new layer of complexity to proteins containing RNA recognition motifs.
Nucleic Acids Res., 42, 2014
|
|
7D6R
 
 | | Crystal structure of the Stx2a complexed with MMA betaAla peptide | | Descriptor: | 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, MMA betaAla peptide, Shiga toxin 2 B subunit, ... | | Authors: | Takahashi, M, Tamada, M, Hibino, M, Senda, M, Okuda, A, Miyazawa, A, Senda, T, Nishikawa, K. | | Deposit date: | 2020-10-01 | | Release date: | 2021-04-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification of a peptide motif that potently inhibits two functionally distinct subunits of Shiga toxin.
Commun Biol, 4, 2021
|
|
