4PM0
 
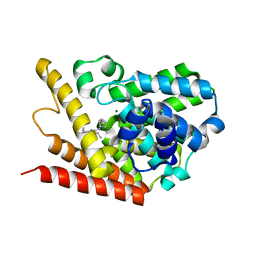 | | PDE7A catalytic domain in complex with 2-(Cyclopentylamino)thieno[3,2-d]pyrimidin-4(3H)-one derivative | | Descriptor: | 2-(cyclopentylamino)-3-ethyl-7-ethynylthieno[3,2-d]pyrimidin-4(3H)-one, High affinity cAMP-specific 3',5'-cyclic phosphodiesterase 7A, MAGNESIUM ION, ... | | Authors: | Kawai, K, Endo, Y, Asano, T, Amano, S, Sawada, K, Ueo, N, Takahashi, N, Sonoda, Y, Kamei, N, Nagata, N. | | Deposit date: | 2014-05-20 | | Release date: | 2014-12-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of 2-(Cyclopentylamino)thieno[3,2-d]pyrimidin-4(3H)-one Derivatives as a New Series of Potent Phosphodiesterase 7 Inhibitors.
J.Med.Chem., 57, 2014
|
|
4Y2B
 
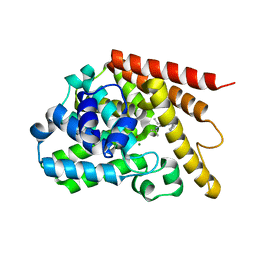 | | Co-crystal structure of 3-ethyl-2-(isopropylamino)-7-(pyridin-3-yl)thieno[3,2-d]pyrimidin-4(3H)-one bound to PDE7A | | Descriptor: | 3-ethyl-2-(propan-2-ylamino)-7-(pyridin-3-yl)thieno[3,2-d]pyrimidin-4(3H)-one, High affinity cAMP-specific 3',5'-cyclic phosphodiesterase 7A, MAGNESIUM ION, ... | | Authors: | Endo, Y, Kawai, K, Asano, T, Amano, S, Asanuma, Y, Sawada, K, Onodera, Y, Ueo, N, Takahashi, N, Sonoda, Y, Kamei, N, Irie, T. | | Deposit date: | 2015-02-09 | | Release date: | 2015-04-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 2-(Isopropylamino)thieno[3,2-d]pyrimidin-4(3H)-one derivatives as selective phosphodiesterase 7 inhibitors with potent in vivo efficacy
Bioorg.Med.Chem.Lett., 25, 2015
|
|
1VDN
 
 | |
4JY2
 
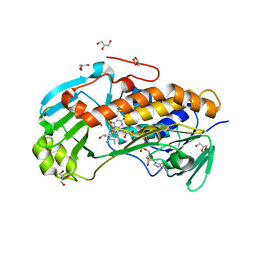 | | Crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, native and unliganded form | | Descriptor: | 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, BETA-MERCAPTOETHANOL, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kobayashi, J, Yoshida, H, Kamitori, S, Hayashi, H, Mizutani, K, Takahashi, N, Mikami, B, Yagi, T. | | Deposit date: | 2013-03-28 | | Release date: | 2014-04-02 | | Method: | X-RAY DIFFRACTION (1.935 Å) | | Cite: | Crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase
To be Published
|
|
4KEQ
 
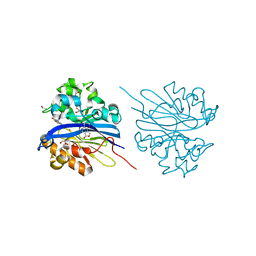 | | Crystal structure of 4-pyridoxolactonase, 5-pyridoxolactone bound | | Descriptor: | 1,2-ETHANEDIOL, 4-pyridoxolactonase, 7-hydroxy-6-methylfuro[3,4-c]pyridin-3(1H)-one, ... | | Authors: | Kobayashi, J, Yoshikane, Y, Baba, S, Mizutani, K, Takahashi, N, Mikami, B, Yagi, T. | | Deposit date: | 2013-04-26 | | Release date: | 2014-04-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.279 Å) | | Cite: | Structure of 4-pyridoxolactonase from Mesorhizobium loti.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
4JY3
 
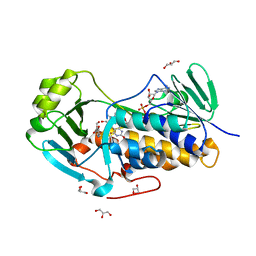 | | Crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, 5-pyridoxic acid bound form | | Descriptor: | 1,2-ETHANEDIOL, 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, 5-hydroxy-4-(hydroxymethyl)-6-methylpyridine-3-carboxylic acid, ... | | Authors: | Kobayashi, J, Yoshida, H, Kamitori, S, Hayashi, H, Mizutani, K, Takahashi, N, Mikami, B, Yagi, T. | | Deposit date: | 2013-03-29 | | Release date: | 2014-04-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase
To be Published
|
|
4KEP
 
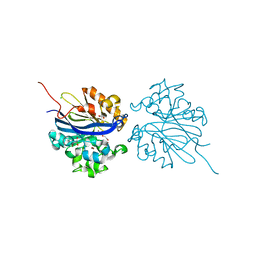 | | Crystal structure of 4-pyridoxolactonase, wild-type | | Descriptor: | 1,2-ETHANEDIOL, 4-pyridoxolactonase, ACETATE ION, ... | | Authors: | Kobayashi, J, Yoshikane, Y, Baba, S, Mizutani, K, Takahashi, N, Mikami, B, Yagi, T. | | Deposit date: | 2013-04-26 | | Release date: | 2014-04-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure of 4-pyridoxolactonase from Mesorhizobium loti.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
8W86
 
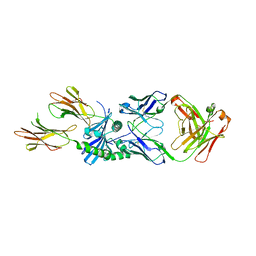 | | HLA-DQ2.5-B/C hordein peptide in complex with DQN0385AE02 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DQN0385AE02 Fab heavy chain, DQN0385AE02 Fab light chain, ... | | Authors: | Irie, M, Tsushima, T, Teranishi-Ikawa, Y, Takahashi, N, Ishii, S, Okura, Y, Fukami, T.A, Torizawa, T. | | Deposit date: | 2023-08-31 | | Release date: | 2023-11-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.236 Å) | | Cite: | Characterizations of a neutralizing antibody broadly reactive to multiple gluten peptide:HLA-DQ2.5 complexes in the context of celiac disease.
Nat Commun, 14, 2023
|
|
8W85
 
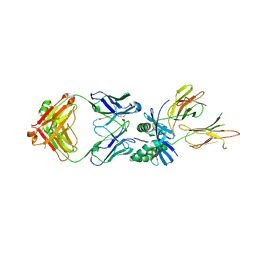 | | HLA-DQ2.5-gamma2 gliadin peptide in complex with DQN0385AE01 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DQN0385AE01 Fab heavy chain, DQN0385AE01 Fab light chain, ... | | Authors: | Irie, M, Tsushima, T, Teranishi-Ikawa, Y, Takahashi, N, Ishii, S, Okura, Y, Fukami, T.A, Torizawa, T. | | Deposit date: | 2023-08-31 | | Release date: | 2023-11-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.769 Å) | | Cite: | Characterizations of a neutralizing antibody broadly reactive to multiple gluten peptide:HLA-DQ2.5 complexes in the context of celiac disease.
Nat Commun, 14, 2023
|
|
8W84
 
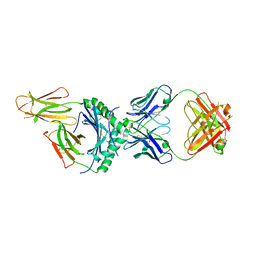 | | HLA-DQ2.5-alpha2 gliadin peptide in complex with DQN0344AE02 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DQN0344AE02 Fab heavy chain, DQN0344AE02 Fab light chain, ... | | Authors: | Irie, M, Tsushima, T, Teranishi-Ikawa, Y, Takahashi, N, Ishii, S, Okura, Y, Fukami, T.A, Torizawa, T. | | Deposit date: | 2023-08-31 | | Release date: | 2023-11-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.105 Å) | | Cite: | Characterizations of a neutralizing antibody broadly reactive to multiple gluten peptide:HLA-DQ2.5 complexes in the context of celiac disease.
Nat Commun, 14, 2023
|
|
8W83
 
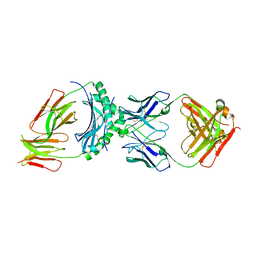 | | HLA-DQ2.5-alpha1 gliadin peptide in complex with DQN0344AE02 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DQN0344AE02 Fab heavy chain, DQN0344AE02 Fab light chain, ... | | Authors: | Irie, M, Tsushima, T, Teranishi-Ikawa, Y, Takahashi, N, Ishii, S, Okura, Y, Fukami, T.A, Torizawa, T. | | Deposit date: | 2023-08-31 | | Release date: | 2023-11-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.818 Å) | | Cite: | Characterizations of a neutralizing antibody broadly reactive to multiple gluten peptide:HLA-DQ2.5 complexes in the context of celiac disease.
Nat Commun, 14, 2023
|
|
6J4H
 
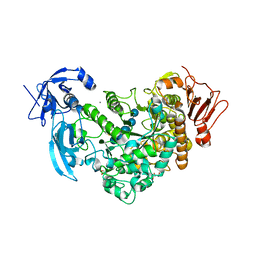 | | Crystal Structure of maltotriose-complex of PulA-G680L mutant from Klebsiella pneumoniae | | Descriptor: | CALCIUM ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Saka, N, Iwamoto, H, Takahashi, N, Mizutani, K, Mikami, B. | | Deposit date: | 2019-01-09 | | Release date: | 2019-09-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.641 Å) | | Cite: | Relationship between the induced-fit loop and the activity of Klebsiella pneumoniae pullulanase.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6J35
 
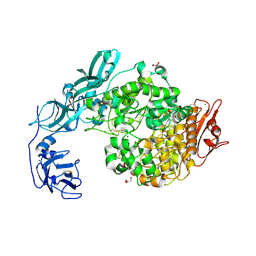 | | Crystal structure of ligand-free of PulA-G680L mutant from Klebsiella pneumoniae | | Descriptor: | CALCIUM ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Saka, N, Iwamoto, H, Takahashi, N, Mizutani, K, Mikami, B. | | Deposit date: | 2019-01-09 | | Release date: | 2019-09-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.839 Å) | | Cite: | Relationship between the induced-fit loop and the activity of Klebsiella pneumoniae pullulanase.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6J33
 
 | | Crystal structure of ligand-free of PulA from Klebsiella pneumoniae | | Descriptor: | ACETATE ION, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Saka, N, Iwamoto, H, Takahashi, N, Mizutani, K, Mikami, B. | | Deposit date: | 2019-01-09 | | Release date: | 2019-09-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.297 Å) | | Cite: | Relationship between the induced-fit loop and the activity of Klebsiella pneumoniae pullulanase.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6J34
 
 | | Crystal Structure of maltotriose-complex of PulA from Klebsiella pneumoniae | | Descriptor: | ACETATE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Saka, N, Iwamoto, H, Takahashi, N, Mizutani, K, Mikami, B. | | Deposit date: | 2019-01-09 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.498 Å) | | Cite: | Relationship between the induced-fit loop and the activity of Klebsiella pneumoniae pullulanase.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
1UHG
 
 | | Crystal Structure of S-Ovalbumin At 1.9 Angstrom Resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ovalbumin, ... | | Authors: | Yamasaki, M, Takahashi, N, Hirose, M. | | Deposit date: | 2003-07-03 | | Release date: | 2003-07-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of S-ovalbumin as a Non-loop-inserted Thermostabilized Serpin Form
J.Biol.Chem., 278, 2003
|
|
5AX3
 
 | | Crystal structure of ERK2 complexed with allosteric and ATP-competitive inhibitors. | | Descriptor: | (2R,3R,4S,5R)-2-(4-AMINO-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL, Mitogen-activated protein kinase 1, allosteric and ATP-competitive inhibitor | | Authors: | Kinoshita, T, Sugiyama, H, Mori, Y, Takahashi, N, Tomonaga, A. | | Deposit date: | 2015-07-14 | | Release date: | 2016-02-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.984 Å) | | Cite: | Identification of allosteric ERK2 inhibitors through in silico biased screening and competitive binding assay
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5YNA
 
 | | Crystal structure of Pullulanase from Klebsiella pneumoniae complex at 1 mM alpha-cyclodextrin | | Descriptor: | ACETATE ION, CALCIUM ION, Cyclohexakis-(1-4)-(alpha-D-glucopyranose), ... | | Authors: | Saka, N, Iwamoto, H, Takahashi, N, Mizutani, K, Mikami, B. | | Deposit date: | 2017-10-24 | | Release date: | 2018-10-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Elucidation of the mechanism of interaction between Klebsiella pneumoniae pullulanase and cyclodextrin
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5YNC
 
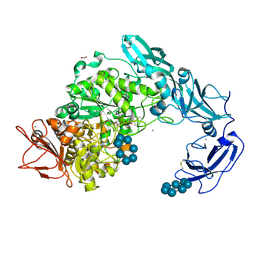 | | Crystal structure of Pullulanase from Klebsiella pneumoniae complex at 1 mM beta-cyclodextrin | | Descriptor: | ACETATE ION, CALCIUM ION, Cycloheptakis-(1-4)-(alpha-D-glucopyranose), ... | | Authors: | Saka, N, Iwamoto, H, Takahashi, N, Mizutani, K, Mikami, B. | | Deposit date: | 2017-10-24 | | Release date: | 2018-10-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Elucidation of the mechanism of interaction between Klebsiella pneumoniae pullulanase and cyclodextrin
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5YNE
 
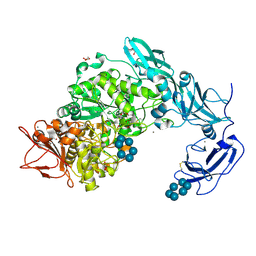 | | Crystal structure of Pullulanase from Klebsiella pneumoniae complex at 10 mM alpha-cyclodextrin | | Descriptor: | CALCIUM ION, Cyclohexakis-(1-4)-(alpha-D-glucopyranose), DI(HYDROXYETHYL)ETHER, ... | | Authors: | Saka, N, Iwamoto, H, Takahashi, N, Mizutani, K, Mikami, B. | | Deposit date: | 2017-10-24 | | Release date: | 2018-10-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Elucidation of the mechanism of interaction between Klebsiella pneumoniae pullulanase and cyclodextrin
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5YN2
 
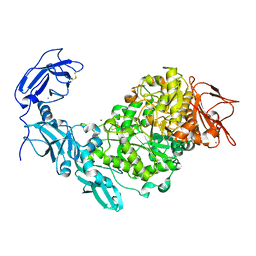 | | Crystal structure of apo Pullulanase from Klebsiella pneumoniae in space group P43212 | | Descriptor: | CALCIUM ION, MAGNESIUM ION, PulA protein | | Authors: | Saka, N, Iwamoto, H, Takahashi, N, Mizutani, K, Mikami, B. | | Deposit date: | 2017-10-24 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Elucidation of the mechanism of interaction between Klebsiella pneumoniae pullulanase and cyclodextrin
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5YN7
 
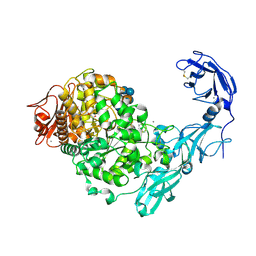 | | Crystal structure of Pullulanase from Klebsiella pneumoniae complex at 0.1 mM beta-cyclodextrin | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Cycloheptakis-(1-4)-(alpha-D-glucopyranose), ... | | Authors: | Saka, N, Iwamoto, H, Takahashi, N, Mizutani, K, Mikami, B. | | Deposit date: | 2017-10-24 | | Release date: | 2018-10-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.586 Å) | | Cite: | Elucidation of the mechanism of interaction between Klebsiella pneumoniae pullulanase and cyclodextrin
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5YND
 
 | | Crystal structure of Pullulanase from Klebsiella pneumoniae complex at 1 mM gamma-cyclodextrin | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, DUF3372 domain-containing protein, ... | | Authors: | Saka, N, Iwamoto, H, Takahashi, N, Mizutani, K, Mikami, B. | | Deposit date: | 2017-10-24 | | Release date: | 2018-10-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.227 Å) | | Cite: | Elucidation of the mechanism of interaction between Klebsiella pneumoniae pullulanase and cyclodextrin
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5YNH
 
 | | Crystal structure of Pullulanase from Klebsiella pneumoniae complex at 10 mM gamma-cyclodextrin | | Descriptor: | CALCIUM ION, Cyclooctakis-(1-4)-(alpha-D-glucopyranose), GLYCEROL, ... | | Authors: | Saka, N, Iwamoto, H, Takahashi, N, Mizutani, K, Mikami, B. | | Deposit date: | 2017-10-24 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Elucidation of the mechanism of interaction between Klebsiella pneumoniae pullulanase and cyclodextrin
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5XBU
 
 | | Crystal structure of GH45 endoglucanase EG27II in apo-form | | Descriptor: | Endo-beta-1,4-glucanase | | Authors: | Nomura, T, Mizutani, K, Iwase, H, Takahashi, N, Mikami, B. | | Deposit date: | 2017-03-21 | | Release date: | 2018-03-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | High-resolution crystal structures of the glycoside hydrolase family 45 endoglucanase EG27II from the snail Ampullaria crossean.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
