4WJ3
 
 | | Crystal structure of the asparagine transamidosome from Pseudomonas aeruginosa | | Descriptor: | 76mer-tRNA, Aspartate--tRNA(Asp/Asn) ligase, Aspartyl/glutamyl-tRNA(Asn/Gln) amidotransferase subunit B, ... | | Authors: | Suzuki, T, Nakamura, A, Kato, K, Tanaka, I, Yao, M. | | Deposit date: | 2014-09-29 | | Release date: | 2014-12-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.705 Å) | | Cite: | Structure of the Pseudomonas aeruginosa transamidosome reveals unique aspects of bacterial tRNA-dependent asparagine biosynthesis
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4WJ4
 
 | | Crystal structure of non-discriminating aspartyl-tRNA synthetase from Pseudomonas aeruginosa complexed with tRNA(Asn) and aspartic acid | | Descriptor: | 76mer-tRNA, ASPARTIC ACID, Aspartate--tRNA(Asp/Asn) ligase | | Authors: | Suzuki, T, Nakamura, A, Kato, K, Tanaka, I, Yao, M. | | Deposit date: | 2014-09-29 | | Release date: | 2014-12-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.294 Å) | | Cite: | Structure of the Pseudomonas aeruginosa transamidosome reveals unique aspects of bacterial tRNA-dependent asparagine biosynthesis
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5UD5
 
 | |
5V6X
 
 | |
6KTQ
 
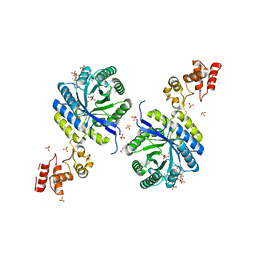 | | Crystal structure of catalytic domain of homocitrate synthase from Sulfolobus acidocaldarius (SaHCS(dRAM)) in complex with alpha-ketoglutarate/Zn2+/CoA | | Descriptor: | 2-OXOGLUTARIC ACID, COENZYME A, Homocitrate synthase, ... | | Authors: | Suzuki, T, Tomita, T, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2019-08-28 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Involvement of subdomain II in the recognition of acetyl-CoA revealed by the crystal structure of homocitrate synthase from Sulfolobus acidocaldarius.
Febs J., 288, 2021
|
|
7CSL
 
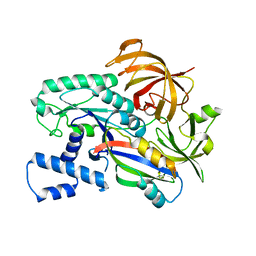 | | Crystal structure of the archaeal EF1A-EF1B complex | | Descriptor: | Elongation factor 1-alpha, Elongation factor 1-beta | | Authors: | Suzuki, T, Ito, K, Miyoshi, T, Murakami, R, Uchiumi, T. | | Deposit date: | 2020-08-15 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the Switching Off of the Interaction between the Archaeal Ribosomal Stalk and aEF1A by Nucleotide Exchange Factor aEF1B.
J.Mol.Biol., 433, 2021
|
|
3SSI
 
 | |
8IB1
 
 | |
7XWA
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Omicron BA.4/5 variant spike protein in complex with its receptor ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Suzuki, T, Kimura, K, Hashiguchi, T. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.36 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 Omicron BA.2 subvariants, including BA.4 and BA.5.
Cell, 185, 2022
|
|
9KKP
 
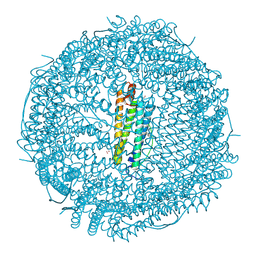 | | Crystal structure of Horse spleen L-ferritin mutant (E53F/E56F/E57F/R59F/E60F/E63F) with Nile Red | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Suzuki, T, Hishikawa, Y, Maity, B, Abe, S, Ueno, T. | | Deposit date: | 2024-11-14 | | Release date: | 2025-03-05 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Design of Aromatic Interaction Networks in a Protein Cage Modulated by Fluorescent Ligand Binding.
Adv Sci, 12, 2025
|
|
9KN7
 
 | | Crystal structure of Horse spleen L-ferritin mutant (Fr-E53F/E56F/E57F/R59L/E60F/E63F) | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Suzuki, T, Hishikawa, Y, Maity, B, Abe, S, Ueno, T. | | Deposit date: | 2024-11-18 | | Release date: | 2025-03-05 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Design of Aromatic Interaction Networks in a Protein Cage Modulated by Fluorescent Ligand Binding.
Adv Sci, 12, 2025
|
|
9KP5
 
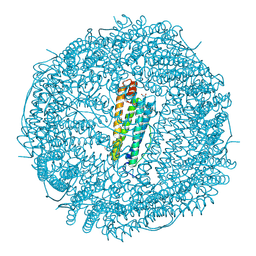 | | Crystal structure of Horse spleen L-ferritin mutant (E56F/R59F) | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Suzuki, T, Hishikawa, Y, Maity, B, Abe, S, Ueno, T. | | Deposit date: | 2024-11-22 | | Release date: | 2025-03-05 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Design of Aromatic Interaction Networks in a Protein Cage Modulated by Fluorescent Ligand Binding.
Adv Sci, 12, 2025
|
|
9KP2
 
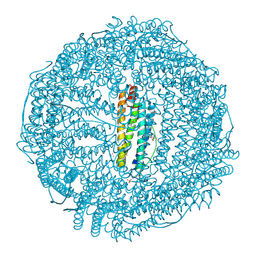 | | Crystal structure of Horse spleen L-ferritin mutant (E56F/R59F) with Nile Red | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Suzuki, T, Hishikawa, Y, Maity, B, Abe, S, Ueno, T. | | Deposit date: | 2024-11-22 | | Release date: | 2025-03-05 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Design of Aromatic Interaction Networks in a Protein Cage Modulated by Fluorescent Ligand Binding.
Adv Sci, 12, 2025
|
|
9KP7
 
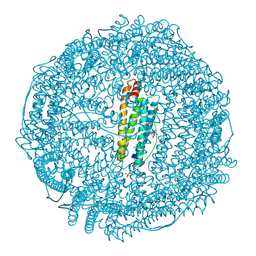 | | Crystal structure of Horse spleen L-ferritin mutant (R59F) with Nile Red | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Suzuki, T, Hishikawa, Y, Maity, B, Abe, S, Ueno, T. | | Deposit date: | 2024-11-22 | | Release date: | 2025-03-05 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Design of Aromatic Interaction Networks in a Protein Cage Modulated by Fluorescent Ligand Binding.
Adv Sci, 12, 2025
|
|
9KPA
 
 | | Crystal structure of Horse spleen L-ferritin mutant (R59F) | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Suzuki, T, Hishikawa, Y, Maity, B, Abe, S, Ueno, T. | | Deposit date: | 2024-11-22 | | Release date: | 2025-03-05 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Design of Aromatic Interaction Networks in a Protein Cage Modulated by Fluorescent Ligand Binding.
Adv Sci, 12, 2025
|
|
9KN8
 
 | | Crystal structure of Horse spleen L-ferritin mutant (E53F/E56F/E57F/R59F/E60F/E63F) with Coumarin 153 | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Suzuki, T, Hishikawa, Y, Maity, B, Abe, S, Ueno, T. | | Deposit date: | 2024-11-18 | | Release date: | 2025-03-05 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Design of Aromatic Interaction Networks in a Protein Cage Modulated by Fluorescent Ligand Binding.
Adv Sci, 12, 2025
|
|
9KLE
 
 | | Crystal structure of Horse spleen L-ferritin mutant (R52F/E56F/R59F/E63F) with Nile Red | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Suzuki, T, Hishikawa, Y, Maity, B, Abe, S, Ueno, T. | | Deposit date: | 2024-11-14 | | Release date: | 2025-03-05 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Design of Aromatic Interaction Networks in a Protein Cage Modulated by Fluorescent Ligand Binding.
Adv Sci, 12, 2025
|
|
9KRS
 
 | | Crystal structure of Horse spleen L-ferritin mutant (Fr-E53F/E56F/E57F/R59A/E60F/E63F) | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Suzuki, T, Hishikawa, Y, Maity, B, Abe, S, Ueno, T. | | Deposit date: | 2024-11-28 | | Release date: | 2025-03-05 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Design of Aromatic Interaction Networks in a Protein Cage Modulated by Fluorescent Ligand Binding.
Adv Sci, 12, 2025
|
|
9KKR
 
 | | Crystal structure of Horse spleen L-ferritin mutant (E53F/E56F/E57F/R59F/E60F/E63F) | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Suzuki, T, Hishikawa, Y, Maity, B, Abe, S, Ueno, T. | | Deposit date: | 2024-11-14 | | Release date: | 2025-03-05 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Design of Aromatic Interaction Networks in a Protein Cage Modulated by Fluorescent Ligand Binding.
Adv Sci, 12, 2025
|
|
3WLU
 
 | | Crystal Structure of human galectin-9 NCRD with Selenolactose | | Descriptor: | 2-(trimethylsilyl)ethyl 4-O-beta-D-galactopyranosyl-6-Se-methyl-6-seleno-beta-D-glucopyranoside, Galectin-9 | | Authors: | Makyio, H, Suzuki, T, Ando, H, Yamada, Y, Ishida, H, Kiso, M, Wakatsuki, S, Kato, R. | | Deposit date: | 2013-11-14 | | Release date: | 2014-11-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Expanded potential of seleno-carbohydrates as a molecular tool for X-ray structural determination of a carbohydrate-protein complex with single/multi-wavelength anomalous dispersion phasing
Bioorg.Med.Chem., 22, 2014
|
|
8HSP
 
 | | E. coli 70S ribosome complexed with tRNA_Ile2 bearing L34 and t6A37 in classical state | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Akiyama, N, Ishiguro, K, Yokoyama, T, Shirouzu, M, Suzuki, T. | | Deposit date: | 2022-12-20 | | Release date: | 2024-04-03 | | Last modified: | 2025-02-12 | | Method: | ELECTRON MICROSCOPY (2.32 Å) | | Cite: | Structural insights into the decoding capability of isoleucine tRNAs with lysidine and agmatidine.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8HU1
 
 | | E. coli 70S ribosome complexed with tRNA_Ile2 bearing L34 and ct6A37 in classical state | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Akiyama, N, Ishiguro, K, Yokoyama, T, Shirouzu, M, Suzuki, T. | | Deposit date: | 2022-12-22 | | Release date: | 2024-04-03 | | Last modified: | 2025-02-12 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Structural insights into the decoding capability of isoleucine tRNAs with lysidine and agmatidine.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8HTZ
 
 | | E. coli 70S ribosome complexed with H. marismortui tRNA_Ile2 bearing agm2C34 in classical state | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Akiyama, N, Ishiguro, K, Yokoyama, T, Shirouzu, M, Suzuki, T. | | Deposit date: | 2022-12-22 | | Release date: | 2024-04-03 | | Last modified: | 2025-02-12 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structural insights into the decoding capability of isoleucine tRNAs with lysidine and agmatidine.
Nat.Struct.Mol.Biol., 31, 2024
|
|
6L3R
 
 | | Crystal structure of Ribonucleotide reductase R1 subunit, RRM1 in complex with 4-bromo-N-((1S,2R)-2-(naphthalen-1-yl)-1-(5-oxo-4,5-dihydro-1,3,4-oxadiazol-2-yl)propyl)benzenesulfonamide | | Descriptor: | 4-bromo-N-((1S,2R)-2-(naphthalen-1-yl)-1-(5-oxo-4,5-dihydro-1,3,4-oxadiazol-2-yl)propyl)benzenesulfonamide, ACETATE ION, MAGNESIUM ION, ... | | Authors: | Miyahara, S, Chong, K.T, Suzuki, T. | | Deposit date: | 2019-10-15 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | TAS1553, a novel small molecule ribonucleotide reductase (RNR) subunit interaction inhibitor, displays remarkable anti-tumor activity
To be published
|
|
6L7L
 
 | | Crystal structure of Ribonucleotide reductase R1 subunit, RRM1 in complex with 5-chloro-2-(N-((1S,2R)-2-(2,3-dihydro-1H-inden-4-yl)-1-(5-oxo-4,5-dihydro-1,3,4-oxadiazol-2-yl)propyl)sulfamoyl)benzamide | | Descriptor: | 5-chloro-2-(N-((1S,2R)-2-(2,3-dihydro-1H-inden-4-yl)-1-(5-oxo-4,5-dihydro-1,3,4-oxadiazol-2-yl)propyl)sulfamoyl)benzamide, ACETATE ION, MAGNESIUM ION, ... | | Authors: | Miyahara, S, Chong, K.T, Suzuki, T. | | Deposit date: | 2019-11-01 | | Release date: | 2020-11-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.171 Å) | | Cite: | TAS1553, a novel small molecule ribonucleotide reductase (RNR) subunit interaction inhibitor, displays remarkable anti-tumor activity
To be published
|
|
