1QZR
 
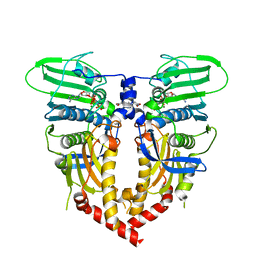 | | CRYSTAL STRUCTURE OF THE ATPASE REGION OF SACCHAROMYCES CEREVISIAE TOPOISOMERASE II BOUND TO ICRF-187 (DEXRAZOXANE) | | Descriptor: | (S)-4,4'-(1-METHYL-1,2-ETHANEDIYL)BIS-2,6-PIPERAZINEDIONE, DNA topoisomerase II, MAGNESIUM ION, ... | | Authors: | Classen, S, Olland, S, Berger, J.M. | | Deposit date: | 2003-09-17 | | Release date: | 2003-09-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the topoisomerase II ATPase region and its mechanism of inhibition by the chemotherapeutic agent ICRF-187
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
2PG6
 
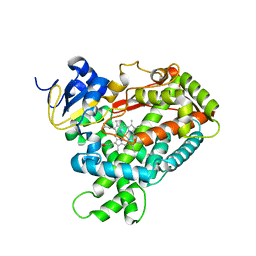 | | Crystal Structure of Human Microsomal P450 2A6 L240C/N297Q | | Descriptor: | Cytochrome P450 2A6, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sansen, S, Hsu, M.H, Stout, C.D, Johnson, E.F. | | Deposit date: | 2007-04-06 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structural insight into the altered substrate specificity of human cytochrome P450 2A6 mutants.
Arch.Biochem.Biophys., 464, 2007
|
|
1RW4
 
 | | Nitrogenase Fe protein l127 deletion variant | | Descriptor: | GLYCEROL, IRON/SULFUR CLUSTER, Nitrogenase iron protein 1 | | Authors: | Sen, S, Igarashi, R, Smith, A, Johnson, M.K, Seefeldt, L.C, Peters, J.W. | | Deposit date: | 2003-12-15 | | Release date: | 2004-03-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Conformational Mimic of the MgATP-Bound "On State" of the Nitrogenase Iron Protein.
Biochemistry, 43, 2004
|
|
2C8V
 
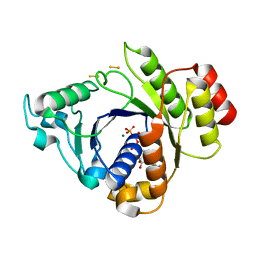 | | Insights into the role of nucleotide-dependent conformational change in nitrogenase catalysis: Structural characterization of the nitrogenase Fe protein Leu127 deletion variant with bound MgATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, FE2/S2 (INORGANIC) CLUSTER, MAGNESIUM ION, ... | | Authors: | Sen, S, Krishnakumar, A, McClead, J, Johnson, M.K, Seefeldt, L.C, Szilagyi, R.K, Peters, J.W. | | Deposit date: | 2005-12-08 | | Release date: | 2006-06-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights Into the Role of Nucleotide-Dependent Conformational Change in Nitrogenase Catalysis: Structural Characterization of the Nitrogenase Fe Protein Leu127 Deletion Variant with Bound Mgatp.
J.Inorg.Biochem., 100, 2006
|
|
4UN2
 
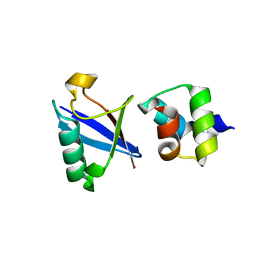 | | Crystal structure of the UBA domain of Dsk2 in complex with Ubiquitin | | Descriptor: | UBIQUITIN, UBIQUITIN DOMAIN-CONTAINING PROTEIN DSK2 | | Authors: | Michielssens, S, Peters, J.H, Ban, D, Pratihar, S, Seeliger, D, Sharma, M, Giller, K, Sabo, T.M, Becker, S, Lee, D, Griesinger, C, de Groot, B.L. | | Deposit date: | 2014-05-23 | | Release date: | 2014-08-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | A Designed Conformational Shift to Control Protein Binding Specificity.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
6Y9T
 
 | | Family GH13_31 enzyme | | Descriptor: | Alpha-glucosidase, CALCIUM ION | | Authors: | Andersen, S, Poulsen, J.C.N, Moeller, M.S, Abou Hachem, M, Lo Leggio, L. | | Deposit date: | 2020-03-10 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | An 1,4-alpha-Glucosyltransferase Defines a New Maltodextrin Catabolism Scheme in Lactobacillus acidophilus.
Appl.Environ.Microbiol., 86, 2020
|
|
4Z1O
 
 | |
4ZFN
 
 | |
8C8G
 
 | |
5BQO
 
 | |
5BQP
 
 | |
7NAM
 
 | | LRP6_E1 in complex with Lr-EET-3.5 | | Descriptor: | Low-density lipoprotein receptor-related protein 6, SODIUM ION, Trypsin inhibitor 2, ... | | Authors: | Hansen, S, Hannoush, R.N. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Directed evolution identifies high-affinity cystine-knot peptide agonists and antagonists of Wnt/ beta-catenin signaling.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6RIM
 
 | |
4DY3
 
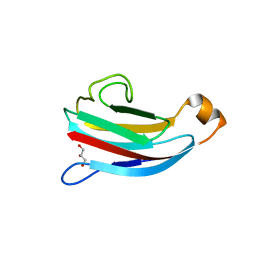 | | crystal structure of Escherichia coli PliG, a periplasmic lysozyme inhibitor of g-type lysozyme | | Descriptor: | GLYCEROL, Inhibitor of g-type lysozyme | | Authors: | Leysen, S, Vanheuverzwijn, S, Van Asten, K, Vanderkelen, L, Michiels, C.W, Strelkov, S.V. | | Deposit date: | 2012-02-28 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural characterization of the PliG lysozyme inhibitor family.
J.Struct.Biol., 180, 2012
|
|
4DY5
 
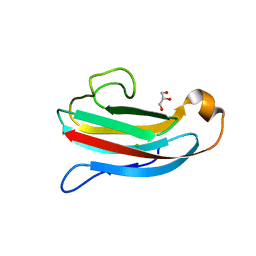 | | Crystal structure of Salmonella Typhimurium PliG, a periplasmic lysozyme inhibitor of g-type lysozyme | | Descriptor: | CHLORIDE ION, GLYCEROL, Gifsy-1 prophage protein | | Authors: | Leysen, S, Theuwis, V, Vanderkelen, L, Michiels, C.W, Strelkov, S.V. | | Deposit date: | 2012-02-28 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural characterization of the PliG lysozyme inhibitor family.
J.Struct.Biol., 180, 2012
|
|
5MA8
 
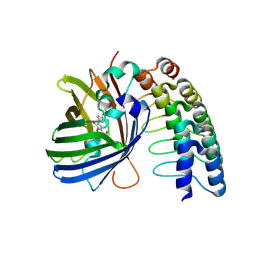 | | GFP-binding DARPin 3G124nc | | Descriptor: | GA-binding protein subunit beta-1, Green fluorescent protein | | Authors: | Hansen, S, Stueber, J, Ernst, P, Koch, A, Bojar, D, Batyuk, A, Plueckthun, A. | | Deposit date: | 2016-11-03 | | Release date: | 2017-12-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Design and applications of a clamp for Green Fluorescent Protein with picomolar affinity.
Sci Rep, 7, 2017
|
|
5MFJ
 
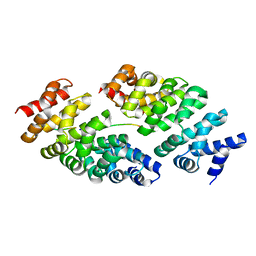 | | Designed armadillo repeat protein YIII(Dq.V2)4CqI in complex with peptide (KR)5 | | Descriptor: | (KR)5, YIII(Dq.V2)4CqI | | Authors: | Hansen, S, Ernst, P, Reichen, C, Ewald, C, Mittl, P, Plueckthun, A. | | Deposit date: | 2016-11-18 | | Release date: | 2017-09-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Curvature of designed armadillo repeat proteins allows modular peptide binding.
J. Struct. Biol., 201, 2018
|
|
5MFO
 
 | | Designed armadillo repeat protein YIIIM3AIII | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, YIIIM3AIII | | Authors: | Hansen, S, Ernst, P, Reichen, C, Ewald, C, Mittl, P, Plueckthun, A. | | Deposit date: | 2016-11-18 | | Release date: | 2017-09-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Curvature of designed armadillo repeat proteins allows modular peptide binding.
J. Struct. Biol., 201, 2018
|
|
5MA6
 
 | | GFP-binding DARPin 3G124nc | | Descriptor: | 1,2-ETHANEDIOL, 3G124nc, Green fluorescent protein, ... | | Authors: | Hansen, S, Stueber, J, Ernst, P, Koch, A, Bojar, D, Batyuk, A, Plueckthun, A. | | Deposit date: | 2016-11-03 | | Release date: | 2017-12-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design and applications of a clamp for Green Fluorescent Protein with picomolar affinity.
Sci Rep, 7, 2017
|
|
5MAK
 
 | | GFP-binding DARPin fusion gc_R7 | | Descriptor: | CITRIC ACID, Green fluorescent protein, R7 | | Authors: | Hansen, S, Stueber, J, Ernst, P, Koch, A, Bojar, D, Batyuk, A, Plueckthun, A. | | Deposit date: | 2016-11-03 | | Release date: | 2017-11-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design and applications of a clamp for Green Fluorescent Protein with picomolar affinity.
Sci Rep, 7, 2017
|
|
5MA4
 
 | | GFP-binding DARPin fusion gc_K7 | | Descriptor: | Green fluorescent protein, K7 | | Authors: | Hansen, S, Stueber, J, Ernst, P, Koch, A, Bojar, D, Batyuk, A, Plueckthun, A. | | Deposit date: | 2016-11-03 | | Release date: | 2017-11-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Design and applications of a clamp for Green Fluorescent Protein with picomolar affinity.
Sci Rep, 7, 2017
|
|
5MFD
 
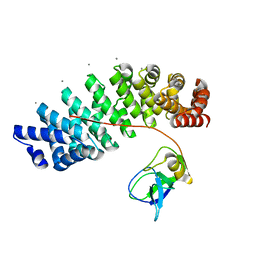 | | Designed armadillo repeat protein YIIIM''6AII in complex with pD_(KR)5 | | Descriptor: | CALCIUM ION, Capsid decoration protein,pD_(KR)5, YIIIM''6AII | | Authors: | Hansen, S, Kiefer, J, Madhurantakam, C, Mittl, P, Plueckthun, A. | | Deposit date: | 2016-11-18 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of designed armadillo repeat proteins binding to peptides fused to globular domains.
Protein Sci., 26, 2017
|
|
5MFC
 
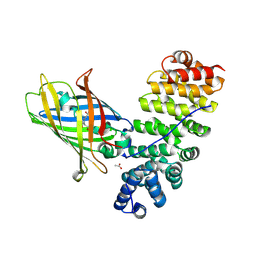 | | Designed armadillo repeat protein YIIIM5AII in complex with (KR)4-GFP | | Descriptor: | (KR)4-Green fluorescent protein,Green fluorescent protein, ACETATE ION, YIIIM5AII | | Authors: | Hansen, S, Kiefer, J, Madhurantakam, C, Mittl, P, Plueckthun, A. | | Deposit date: | 2016-11-18 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of designed armadillo repeat proteins binding to peptides fused to globular domains.
Protein Sci., 26, 2017
|
|
5MFL
 
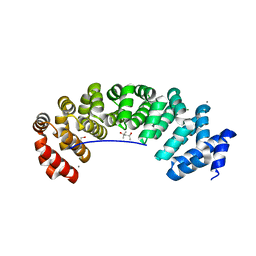 | | Designed armadillo repeat protein (KR)5_GS10_YIIIM6AII | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (KR)5_GS10_YIIIM6AII, 1,2-ETHANEDIOL, ... | | Authors: | Hansen, S, Ernst, P, Reichen, C, Ewald, C, Mittl, P, Plueckthun, A. | | Deposit date: | 2016-11-18 | | Release date: | 2017-09-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Curvature of designed armadillo repeat proteins allows modular peptide binding.
J. Struct. Biol., 201, 2018
|
|
5MFE
 
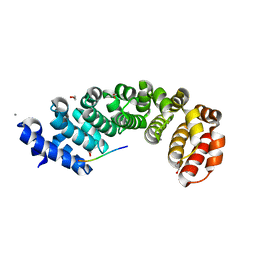 | | Designed armadillo repeat protein YIIIM5AII in complex with (RR)4 peptide | | Descriptor: | (RR)4, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Hansen, S, Ernst, P, Reichen, C, Ewald, C, Mittl, P, Plueckthun, A. | | Deposit date: | 2016-11-18 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Curvature of designed armadillo repeat proteins allows modular peptide binding.
J. Struct. Biol., 201, 2018
|
|
