6Y1A
 
 | | Amyloid fibril structure of islet amyloid polypeptide | | Descriptor: | AMINO GROUP, Islet amyloid polypeptide | | Authors: | Roeder, C, Kupreichyk, T, Gremer, L, Schaefer, L.U, Pothula, K.R, Ravelli, R.B.G, Willbold, D, Hoyer, W, Schroder, G.F. | | Deposit date: | 2020-02-11 | | Release date: | 2020-03-04 | | Last modified: | 2020-07-22 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structure of islet amyloid polypeptide fibrils reveals similarities with amyloid-beta fibrils.
Nat.Struct.Mol.Biol., 27, 2020
|
|
8FEJ
 
 | |
8FEL
 
 | |
5FQ8
 
 | | Crystal structure of the SusCD complex BT2261-2264 from Bacteroides thetaiotaomicron | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 3-decanoyloxypropyl decanoate, BT_2262 (UNCHARACTERISED LIPOPROTEIN), ... | | Authors: | Glenwright, A.J, Pothula, K.R, Chorev, D.S, Basle, A, Robinson, C.V, Kleinekathoefer, U, Bolam, D.N, van den Berg, B. | | Deposit date: | 2015-12-07 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for nutrient acquisition by dominant members of the human gut microbiota.
Nature, 541, 2017
|
|
5FQ6
 
 | | Crystal structure of the SusCD complex BT2261-2264 from Bacteroides thetaiotaomicron | | Descriptor: | 3-decanoyloxypropyl decanoate, BT_2261, CALCIUM ION, ... | | Authors: | Glenwright, A.J, Pothula, K.R, Chorev, D.S, Basle, A, Robinson, C.V, Kleinekathoefer, U, Bolam, D.N, van den Berg, B. | | Deposit date: | 2015-12-07 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for nutrient acquisition by dominant members of the human gut microbiota.
Nature, 541, 2017
|
|
5FQ7
 
 | | Crystal structure of the SusCD complex BT2261-2264 from Bacteroides thetaiotaomicron | | Descriptor: | BT_2261, BT_2262, BT_2263, ... | | Authors: | Glenwright, A.J, Pothula, K.R, Chorev, D.S, Basle, A, Robinson, C.V, Kleinekathoefer, U, Bolam, D.N, van den Berg, B. | | Deposit date: | 2015-12-07 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural basis for nutrient acquisition by dominant members of the human gut microbiota.
Nature, 541, 2017
|
|
5FQ4
 
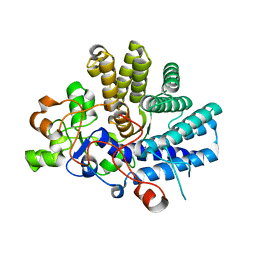 | | Crystal structure of the lipoprotein BT2263 from Bacteroides thetaiotaomicron | | Descriptor: | CALCIUM ION, PUTATIVE LIPOPROTEIN | | Authors: | Glenwright, A.J, Pothula, K.R, Chorev, D.S, Basle, A, Robinson, C.V, Kleinekathoefer, U, Bolam, D.N, van den Berg, B. | | Deposit date: | 2015-12-04 | | Release date: | 2016-12-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for nutrient acquisition by dominant members of the human gut microbiota.
Nature, 541, 2017
|
|
5FQ3
 
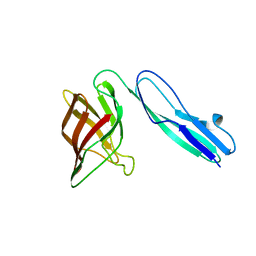 | | Crystal structure of the lipoprotein BT2262 from Bacteroides thetaiotaomicron | | Descriptor: | BT_2262 | | Authors: | Glenwright, A.J, Pothula, K.R, Chorev, D.S, Basle, A, Robinson, C.V, Kleinekathoefer, U, Bolam, D.N, van den Berg, B. | | Deposit date: | 2015-12-04 | | Release date: | 2016-12-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for nutrient acquisition by dominant members of the human gut microbiota.
Nature, 541, 2017
|
|
8TGW
 
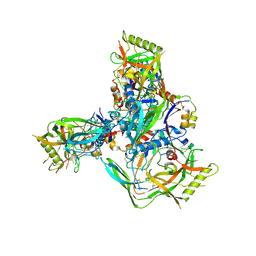 | | Cryo-EM structure of 1059 SOSIP trimer purified via Galanthus nivalis lectin chromatography | | Descriptor: | 1059 SOSIP Surface protein gp120, 1059 SOSIP Transmembrane protein gp41, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Parsons, R.J, Pothula, K, Acharya, P. | | Deposit date: | 2023-07-13 | | Release date: | 2024-08-28 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Conformational flexibility of HIV-1 envelope glycoproteins modulates transmitted/founder sensitivity to broadly neutralizing antibodies.
Nat Commun, 15, 2024
|
|
8TGU
 
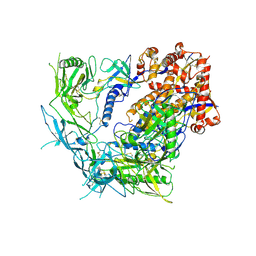 | | Cryo-EM structure of BG505 SOSIP trimer purified via Galanthus nivalis lectin chromatography | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 Envelope glycoprotein BG505 SOSIP, ... | | Authors: | Parsons, R.J, Pothula, K, Acharya, P. | | Deposit date: | 2023-07-13 | | Release date: | 2024-08-28 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Conformational flexibility of HIV-1 envelope glycoproteins modulates transmitted/founder sensitivity to broadly neutralizing antibodies.
Nat Commun, 15, 2024
|
|
5T3R
 
 | | Crystal structure of BT1762-1763 | | Descriptor: | MAGNESIUM ION, SODIUM ION, SusC homolog, ... | | Authors: | van den Berg, B. | | Deposit date: | 2016-08-26 | | Release date: | 2016-12-14 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for nutrient acquisition by dominant members of the human gut microbiota.
Nature, 541, 2017
|
|
5T4Y
 
 | | Crystal structure of BT1762-1763 | | Descriptor: | MAGNESIUM ION, SusC homolog, SusD homolog | | Authors: | van den Berg, B. | | Deposit date: | 2016-08-30 | | Release date: | 2016-12-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for nutrient acquisition by dominant members of the human gut microbiota.
Nature, 541, 2017
|
|
7PDC
 
 | |
5MDP
 
 | |
5MDS
 
 | |
5MDO
 
 | |
5MDQ
 
 | |
5LX8
 
 | | Crystal structure of BT1762 | | Descriptor: | SODIUM ION, SULFATE ION, SusD homolog | | Authors: | Basle, A. | | Deposit date: | 2016-09-20 | | Release date: | 2016-12-14 | | Last modified: | 2017-08-30 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural basis for nutrient acquisition by dominant members of the human gut microbiota.
Nature, 541, 2017
|
|
5MDR
 
 | | Crystal structure of in vitro folded Chitoporin VhChip from Vibrio harveyi in complex with chitohexaose | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitoporin, ... | | Authors: | Zahn, M, van den Berg, B. | | Deposit date: | 2016-11-13 | | Release date: | 2017-12-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for chitin acquisition by marine Vibrio species.
Nat Commun, 9, 2018
|
|
