2DEA
 
 | |
3YPI
 
 | |
6FAB
 
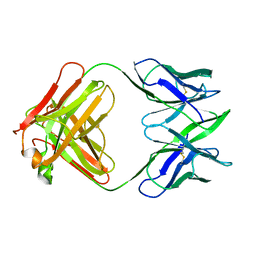 | | THREE-DIMENSIONAL STRUCTURE OF MURINE ANTI-P-AZOPHENYLARSONATE FAB 36-71. 1. X-RAY CRYSTALLOGRAPHY, SITE-DIRECTED MUTAGENESIS, AND MODELING OF THE COMPLEX WITH HAPTEN | | Descriptor: | IGG1-KAPPA 36-71 FAB (HEAVY CHAIN), IGG1-KAPPA 36-71 FAB (LIGHT CHAIN) | | Authors: | Strong, R.K, Rose, D.R, Petsko, G.A, Sharon, J, Margolies, M.N. | | Deposit date: | 1991-01-17 | | Release date: | 1993-01-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Three-dimensional structure of murine anti-p-azophenylarsonate Fab 36-71. 1. X-ray crystallography, site-directed mutagenesis, and modeling of the complex with hapten.
Biochemistry, 30, 1991
|
|
1TXR
 
 | | X-ray crystal structure of bestatin bound to AAP | | Descriptor: | 2-(3-AMINO-2-HYDROXY-4-PHENYL-BUTYRYLAMINO)-4-METHYL-PENTANOIC ACID, Bacterial leucyl aminopeptidase, ZINC ION | | Authors: | Stamper, C.C, Holz, R.C, Ringe, D, Petsko, G.A. | | Deposit date: | 2004-07-06 | | Release date: | 2004-07-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Spectroscopic and X-ray Crystallographic Characterization of Bestatin Bound to the Aminopeptidase from Aeromonas (Vibrio) proteolytica.
Biochemistry, 43, 2004
|
|
2YPI
 
 | |
9RAT
 
 | |
2MNR
 
 | |
3SDP
 
 | |
3FZW
 
 | | Crystal Structure of Ketosteroid Isomerase D40N-D103N from Pseudomonas putida (pKSI) with bound equilenin | | Descriptor: | EQUILENIN, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Caaveiro, J.M.M, Ringe, D, Petsko, G.A. | | Deposit date: | 2009-01-26 | | Release date: | 2009-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Hydrogen bond coupling in the ketosteroid isomerase active site.
Biochemistry, 48, 2009
|
|
7RAT
 
 | |
5GCH
 
 | |
2INX
 
 | | Crystal Structure of Ketosteroid Isomerase D40N from Pseudomonas putida (pKSI) with bound 2,6-difluorophenol | | Descriptor: | 2,6-DIFLUOROPHENOL, Steroid delta-isomerase | | Authors: | Martinez Caaveiro, J.M, Pybus, B, Ringe, D, Petsko, G.A, Sigala, P, Kraut, D, Herschlag, D. | | Deposit date: | 2006-10-09 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Testing geometrical discrimination within an enzyme active site: constrained hydrogen bonding in the ketosteroid isomerase oxyanion hole.
J.Am.Chem.Soc., 130, 2008
|
|
4GCH
 
 | |
3LZ2
 
 | | STRUCTURE DETERMINATION OF TURKEY EGG WHITE LYSOZYME USING LAUE DIFFRACTION | | Descriptor: | TURKEY EGG WHITE LYSOZYME | | Authors: | Howell, P.L, Almo, S.C, Parsons, M.R, Hajdu, J, Petsko, G.A. | | Deposit date: | 1991-09-13 | | Release date: | 1993-10-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure determination of turkey egg-white lysozyme using Laue diffraction data.
Acta Crystallogr.,Sect.B, 48, 1992
|
|
7UGR
 
 | | Crystal structure of hyperfolder YFP | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Hyperfolder yellow fluorescent protein, ... | | Authors: | Campbell, B.C, Liu, C.F, Petsko, G.A. | | Deposit date: | 2022-03-25 | | Release date: | 2022-10-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Chemically stable fluorescent proteins for advanced microscopy.
Nat.Methods, 19, 2022
|
|
7UGS
 
 | |
7UGT
 
 | |
6C2Z
 
 | | Crystal Structures of Cystathionine beta-Synthase from Saccharomyces cerevisiae: the Structure of the PLP-Aminoacrylate Intermediate | | Descriptor: | 1,2-ETHANEDIOL, 2-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]ACRYLIC ACID, CALCIUM ION, ... | | Authors: | Kreinbring, C.A, Tu, Y, Liu, D, Petsko, G.A, Ringe, D. | | Deposit date: | 2018-01-09 | | Release date: | 2018-04-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Crystal Structures of Cystathionine beta-Synthase from Saccharomyces cerevisiae: One Enzymatic Step at a Time.
Biochemistry, 57, 2018
|
|
3DHC
 
 | | 1.3 Angstrom Structure of N-Acyl Homoserine Lactone Hydrolase with the Product N-Hexanoyl-L-Homocysteine Bound to The catalytic Metal Center | | Descriptor: | GLYCEROL, N-Acyl Homoserine Lactone Hydrolase, N-hexanoyl-L-homocysteine, ... | | Authors: | Liu, D, Momb, J, Thomas, P.W, Moulin, A, Petsko, G.A, Fast, W, Ringe, D. | | Deposit date: | 2008-06-17 | | Release date: | 2008-07-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Mechanism of the quorum-quenching lactonase (AiiA) from Bacillus thuringiensis. 1. Product-bound structures.
Biochemistry, 47, 2008
|
|
3DHA
 
 | | An Ultral High Resolution Structure of N-Acyl Homoserine Lactone Hydrolase with the Product N-Hexanoyl-L-Homoserine Bound at An Alternative Site | | Descriptor: | GLYCEROL, N-Acyl Homoserine Lactone Hydrolase, N-hexanoyl-L-homoserine, ... | | Authors: | Liu, D, Momb, J, Thomas, P.W, Moulin, A, Petsko, G.A, Fast, W, Ringe, D. | | Deposit date: | 2008-06-17 | | Release date: | 2008-07-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Mechanism of the quorum-quenching lactonase (AiiA) from Bacillus thuringiensis. 1. Product-bound structures.
Biochemistry, 47, 2008
|
|
3DHB
 
 | | 1.4 Angstrom Structure of N-Acyl Homoserine Lactone Hydrolase with the Product N-Hexanoyl-L-Homoserine Bound at The Catalytic Metal Center | | Descriptor: | GLYCEROL, N-Acyl Homoserine Lactone Hydrolase, N-hexanoyl-L-homoserine, ... | | Authors: | Liu, D, Momb, J, Thomas, P.W, Moulin, A, Petsko, G.A, Fast, W, Ringe, D. | | Deposit date: | 2008-06-17 | | Release date: | 2008-07-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Mechanism of the quorum-quenching lactonase (AiiA) from Bacillus thuringiensis. 1. Product-bound structures.
Biochemistry, 47, 2008
|
|
2SFP
 
 | | ALANINE RACEMASE WITH BOUND PROPIONATE INHIBITOR | | Descriptor: | PROPANOIC ACID, PROTEIN (ALANINE RACEMASE), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Morollo, A.A, Petsko, G.A, Ringe, D. | | Deposit date: | 1999-02-16 | | Release date: | 1999-02-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a Michaelis complex analogue: propionate binds in the substrate carboxylate site of alanine racemase.
Biochemistry, 38, 1999
|
|
8RAT
 
 | |
6RHN
 
 | | HISTIDINE TRIAD NUCLEOTIDE-BINDING PROTEIN (HINT) FROM RABBIT WITHOUT NUCLEOTIDE | | Descriptor: | HISTIDINE TRIAD NUCLEOTIDE-BINDING PROTEIN | | Authors: | Brenner, C, Garrison, P, Gilmour, J, Peisach, D, Ringe, D, Petsko, G.A, Lowenstein, J.M. | | Deposit date: | 1997-02-27 | | Release date: | 1997-06-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structures of HINT demonstrate that histidine triad proteins are GalT-related nucleotide-binding proteins.
Nat.Struct.Biol., 4, 1997
|
|
7M7C
 
 | | Crystal Structure of Hip1 (Rv2224c) mutant - T466A/S228DHA (dehydroalanine) | | Descriptor: | Carboxylesterase A | | Authors: | Naffin-Olivos, J.L, Daab, A, Goldfarb, N.E, Doran, M.H, Baikovitz, J, Liu, D, Sun, S, White, A, Dunn, B.M, Rengarajan, J, Petsko, G.A, Ringe, D. | | Deposit date: | 2021-03-27 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Inhibitors and Inactivators of Mycobacterium tuberculosis serine protease Hip1 (Rv2224c)
To Be Published
|
|
