1US3
 
 | | Native xylanase10C from Cellvibrio japonicus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ENDO-BETA-1,4-XYLANASE PRECURSOR, GLYCEROL, ... | | Authors: | Pell, G, Szabo, L, Charnock, S.J, Xie, H, Gloster, T.M, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2003-11-17 | | Release date: | 2003-12-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Biochemical Analysis of Cellvibrio Japonicus Xylanase 10C: How Variation in Substrate-Binding Cleft Influences the Catalytic Profile of Family Gh-10 Xylanases
J.Biol.Chem., 279, 2004
|
|
1UQZ
 
 | | Xylanase Xyn10B mutant (E262S) from Cellvibrio mixtus in complex with 4-O-methyl glucuronic acid | | Descriptor: | 4-O-methyl-alpha-D-glucopyranuronic acid-(1-2)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, CHLORIDE ION, ENDOXYLANASE, ... | | Authors: | Pell, G, Taylor, E.J, Gloster, T.M, Turkenburg, J.P, Fontes, C.M.G.A, Ferreira, L.M.A, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2003-10-24 | | Release date: | 2003-12-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Mechanisms by which Family 10 Glycoside Hydrolases Bind Decorated Substrates
J.Biol.Chem., 279, 2004
|
|
1UR2
 
 | | Xylanase Xyn10B mutant (E262S) from Cellvibrio mixtus in complex with arabinofuranose alpha 1,3 linked to xylotriose | | Descriptor: | CHLORIDE ION, ENDOXYLANASE, MAGNESIUM ION, ... | | Authors: | Pell, G, Taylor, E.J, Gloster, T.M, Turkenburg, J.P, Fontes, C.M.G.A, Ferreira, L.M.A, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2003-10-24 | | Release date: | 2003-12-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Mechanisms by which Family 10 Glycoside Hydrolases Bind Decorated Substrates
J.Biol.Chem., 279, 2004
|
|
1UQY
 
 | | Xylanase Xyn10B mutant (E262S) from Cellvibrio mixtus in complex with xylopentaose | | Descriptor: | ENDOXYLANASE, MAGNESIUM ION, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-alpha-D-xylopyranose, ... | | Authors: | Pell, G, Taylor, E.J, Gloster, T.M, Turkenburg, J.P, Fontes, C.M.G.A, Ferreira, L.M.A, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2003-10-23 | | Release date: | 2003-12-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | The Mechanisms by which Family 10 Glycoside Hydrolases Bind Decorated Substrates
J.Biol.Chem., 279, 2004
|
|
1UR1
 
 | | Xylanase Xyn10B mutant (E262S) from Cellvibrio mixtus in complex with arabinofuranose alpha-1,3 linked to xylobiose | | Descriptor: | CHLORIDE ION, ENDOXYLANASE, MAGNESIUM ION, ... | | Authors: | Pell, G, Taylor, E.J, Gloster, T.M, Turkenburg, J.P, Fontes, C.M.G.A, Ferreira, L.M.A, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2003-10-24 | | Release date: | 2003-12-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | The Mechanisms by which Family 10 Glycoside Hydrolases Bind Decorated Substrates
J.Biol.Chem., 279, 2004
|
|
1US2
 
 | | Xylanase10C (mutant E385A) from Cellvibrio japonicus in complex with xylopentaose | | Descriptor: | ENDO-BETA-1,4-XYLANASE, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Pell, G, Szabo, L, Charnock, S.J, Xie, H, Gloster, T.M, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2003-11-17 | | Release date: | 2003-12-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Biochemical Analysis of Cellvibrio Japonicus Xylanase 10C: How Variation in Substrate-Binding Cleft Influences the Catalytic Profile of Family Gh-10 Xylanases
J.Biol.Chem., 279, 2004
|
|
2BGO
 
 | | Mannan Binding Module from Man5C | | Descriptor: | ENDO-B1,4-MANNANASE 5C | | Authors: | Tunnicliffe, R.B, Bolam, D.N, Pell, G, Gilbert, H.J, Williamson, M.P. | | Deposit date: | 2005-01-04 | | Release date: | 2005-03-09 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure of a Mannan-Specific Family 35 Carbohydrate-Binding Module: Evidence for Significant Conformational Changes Upon Ligand Binding
J.Mol.Biol., 347, 2005
|
|
2BGP
 
 | | Mannan Binding Module from Man5C in bound conformation | | Descriptor: | ENDO-B1,4-MANNANASE 5C | | Authors: | Tunnicliffe, R.B, Bolam, D.N, Pell, G, Gilbert, H.J, Williamson, M.P. | | Deposit date: | 2005-01-04 | | Release date: | 2005-03-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of a Mannan-Specific Family 35 Carbohydrate-Binding Module: Evidence for Significant Conformational Changes Upon Ligand Binding
J.Mol.Biol., 347, 2005
|
|
1OCN
 
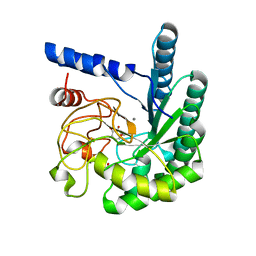 | | Mutant D416A of the CELLOBIOHYDROLASE CEL6A FROM HUMICOLA INSOLENS in complex with a cellobio-derived isofagomine at 1.3 angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, CALCIUM ION, ... | | Authors: | Varrot, A, Macdonald, J, Stick, R.V, Pell, G, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2003-02-09 | | Release date: | 2003-05-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Distortion of a Cellobio-Derived Isofagomine Highlights the Potential Conformational Itinerary of Inverting Beta-Glucosidases
Chem.Commun.(Camb.), 21, 2003
|
|
1ODT
 
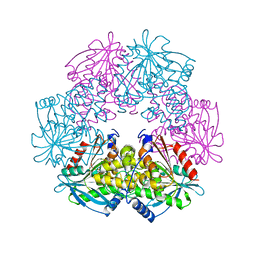 | | cephalosporin C deacetylase mutated, in complex with acetate | | Descriptor: | ACETATE ION, CEPHALOSPORIN C DEACETYLASE | | Authors: | Vincent, F, Charnock, S.J, Verschueren, K.H.G, Turkenburg, J.P, Scott, D.J, Offen, W.A, Roberts, S, Pell, G, Gilbert, H.J, Brannigan, J.A, Davies, G.J. | | Deposit date: | 2003-02-20 | | Release date: | 2003-07-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Multifunctional Xylooligosaccharide/Cephalosporin C Deacetylase Revealed by the Hexameric Structure of the Bacillus Subtilis Enzyme at 1.9A Resolution
J.Mol.Biol., 330, 2003
|
|
1ODS
 
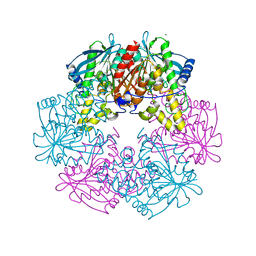 | | Cephalosporin C deacetylase from Bacillus subtilis | | Descriptor: | CEPHALOSPORIN C DEACETYLASE, CHLORIDE ION, MAGNESIUM ION | | Authors: | Vincent, F, Charnock, S.J, Verschueren, K.H.G, Turkenburg, J.P, Scott, D.J, Offen, W.A, Roberts, S, Pell, G, Gilbert, H.J, Brannigan, J.A, Davies, G.J. | | Deposit date: | 2003-02-20 | | Release date: | 2003-07-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Multifunctional Xylooligosaccharide/Cephalosporin C Deacetylase Revealed by the Hexameric Structure of the Bacillus Subtilis Enzyme at 1.9A Resolution
J.Mol.Biol., 330, 2003
|
|
1UP3
 
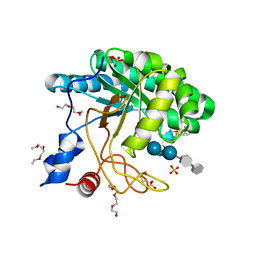 | | Structure of the endoglucanase Cel6 from Mycobacterium tuberculosis in complex with METHYL-CELLOBIOSYL-4-DEOXY-4-THIO-BETA-D-CELLOBIOSIDE at 1.6 angstrom | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, PUTATIVE CELLULASE CEL6, SULFATE ION, ... | | Authors: | Varrot, A, Leydier, S, Pell, G, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2003-09-26 | | Release date: | 2004-11-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mycobacterium Tuberculosis Strains Possess Functional Cellulases.
J.Biol.Chem., 280, 2005
|
|
1UOZ
 
 | | Structure of the endoglucanase Cel6 from Mycobacterium tuberculosis in complex with thiocellopentaose at 1.1 angstrom | | Descriptor: | 4-thio-beta-D-glucopyranose, GLYCEROL, PUTATIVE CELLULASE, ... | | Authors: | Varrot, A, Leydier, S, Pell, G, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2003-09-26 | | Release date: | 2004-11-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Mycobacterium Tuberculosis Strains Possess Functional Cellulases.
J.Biol.Chem., 280, 2005
|
|
1UUQ
 
 | | Exo-mannosidase from Cellvibrio mixtus | | Descriptor: | GLYCEROL, MANNOSYL-OLIGOSACCHARIDE GLUCOSIDASE, SULFATE ION | | Authors: | Dias, M.V.F, Vincent, F, Pell, G, Prates, J.A.M, Centeno, M.S.J, Ferreira, L.M.A, Gilbert, H.J, Davies, G.J, Fontes, C.M.G.A. | | Deposit date: | 2004-01-09 | | Release date: | 2004-04-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Insights Into the Molecular Determinants of Substrate Specificity in Glycoside Hydrolase Family 5 Revealed by the Crystal Structure and Kinetics of Cellvibrio Mixtus Mannosidase 5A
J.Biol.Chem., 279, 2004
|
|
1UP2
 
 | | Structure of the endoglucanase Cel6 from Mycobacterium tuberculosis in complex with glucose-isofagomine at 1.9 angstrom | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, ACETATE ION, ... | | Authors: | Varrot, A, Leydier, S, Pell, G, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2003-09-26 | | Release date: | 2004-11-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mycobacterium Tuberculosis Strains Possess Functional Cellulases.
J.Biol.Chem., 280, 2005
|
|
1UP0
 
 | | Structure of the endoglucanase Cel6 from Mycobacterium tuberculosis in complex with cellobiose at 1.75 angstrom | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, ACETATE ION, PUTATIVE CELLULASE CEL6, ... | | Authors: | Varrot, A, Leydier, S, Pell, G, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2003-09-26 | | Release date: | 2004-11-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Mycobacterium Tuberculosis Strains Possess Functional Cellulases.
J.Biol.Chem., 280, 2005
|
|
3ZOW
 
 | | Crystal Structure of Wild Type Nitrosomonas europaea Cytochrome c552 | | Descriptor: | CYTOCHROME C-552, HEME C | | Authors: | Hersleth, H.-P, Can, M, Krucinska, J, Zoppellaro, G, Andersen, N.H, Karlsen, S, Wedekind, J.E, Andersson, K.K, Bren, K.L. | | Deposit date: | 2013-02-25 | | Release date: | 2013-08-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Characterization of Nitrosomonas Europaea Cytochrome C-552 Variants with Marked Differences in Electronic Structure.
Chembiochem, 14, 2013
|
|
3ZOX
 
 | | Crystal Structure of N64Del Mutant of Nitrosomonas europaea Cytochrome c552 (monoclinic space group) | | Descriptor: | CYTOCHROME C-552, HEME C | | Authors: | Hersleth, H.-P, Can, M, Krucinska, J, Zoppellaro, G, Andersen, N.H, Wedekind, J.E, Andersson, K.K, Bren, K.L. | | Deposit date: | 2013-02-26 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Characterization of Nitrosomonas Europaea Cytochrome C-552 Variants with Marked Differences in Electronic Structure.
Chembiochem, 14, 2013
|
|
8AWW
 
 | | Transthyretin conjugated with a tafamidis derivative | | Descriptor: | Transthyretin, ~{N}-(6-azanylhexyl)-2-[3,5-bis(chloranyl)phenyl]-1,3-benzoxazole-6-carboxamide | | Authors: | Cerofolini, L, Vasa, K, Bianconi, E, Salobehaj, M, Cappelli, G, Licciardi, G, Perez-Rafols, A, Padilla Cortes, L.D, Antonacci, S, Rizzo, D, Ravera, E, Calderone, V, Parigi, G, Luchinat, C, Macchiarulo, A, Menichetti, S, Fragai, M. | | Deposit date: | 2022-08-30 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Combining Solid-State NMR with Structural and Biophysical Techniques to Design Challenging Protein-Drug Conjugates.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
3ZOY
 
 | | Crystal Structure of N64Del Mutant of Nitrosomonas europaea Cytochrome c552 (hexagonal space group) | | Descriptor: | CYTOCHROME C-552, HEME C | | Authors: | Hersleth, H.-P, Can, M, Krucinska, J, Zoppellaro, G, Andersen, N.H, Wedekind, J.E, Andersson, K.K, Bren, K.L. | | Deposit date: | 2013-02-26 | | Release date: | 2013-08-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Characterization of Nitrosomonas Europaea Cytochrome C-552 Variants with Marked Differences in Electronic Structure.
Chembiochem, 14, 2013
|
|
5LP6
 
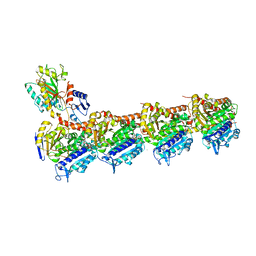 | | Crystal structure of Tubulin-Stathmin-TTL-Thiocolchicine Complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Marangon, J, Christodoulou, M, Casagrande, F, Tiana, G, Dalla Via, L, Aliverti, A, Passarella, D, Cappelletti, G, Ricagno, S. | | Deposit date: | 2016-08-11 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Tools for the rational design of bivalent microtubule-targeting drugs.
Biochem. Biophys. Res. Commun., 479, 2016
|
|
6BBL
 
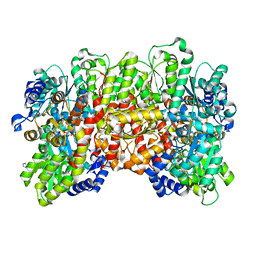 | | Crystal structure of the a-96Gln MoFe protein variant in the presence of the substrate acetylene | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, FE (III) ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Zadvornyy, O.A, Keable, S.M, Vertemara, J, Eilers, B.J, Karamatullah, D, Rasmussen, A.J, De Gioia, L, Zampella, G, Seefeldt, L.C, Peters, J.W. | | Deposit date: | 2017-10-18 | | Release date: | 2018-01-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural characterization of the nitrogenase molybdenum-iron protein with the substrate acetylene trapped near the active site.
J. Inorg. Biochem., 180, 2017
|
|
1W2P
 
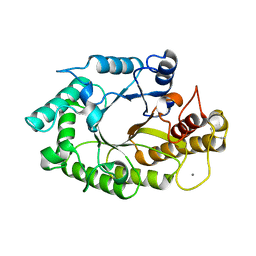 | | The 3-dimensional structure of a xylanase (Xyn10A) from Cellvibrio japonicus | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, ENDO-1,4-BETA-XYLANASE A PRECURSOR | | Authors: | Taylor, E.J, Vincent, F, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2004-07-07 | | Release date: | 2004-09-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Use of Forced Protein Evolution to Investigate and Improve Stability of Family 10 Xylanases: The Production of Ca2+-Independent Stable Xylanases
J.Biol.Chem., 279, 2004
|
|
4JCG
 
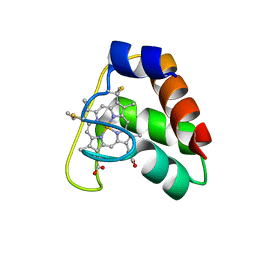 | | Recombinant wild type Nitrosomonas europaea cytochrome c552 | | Descriptor: | Cytochrome c-552, HEME C | | Authors: | Wedekind, J.E, Can, M, Krucinska, J, Bren, K.L. | | Deposit date: | 2013-02-21 | | Release date: | 2013-08-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural Characterization of Nitrosomonas europaea Cytochrome c-552 Variants with Marked Differences in Electronic Structure.
Chembiochem, 14, 2013
|
|
