2GUI
 
 | | Structure and Function of Cyclized Versions of the Proofreading Exonuclease Subunit of E. coli DNA Polymerase III | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, DNA polymerase III epsilon subunit, ... | | Authors: | Park, A.Y, Carr, P.D, Ollis, D.L, Dixon, N.E. | | Deposit date: | 2006-04-30 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and Function of Cyclized Versions of the Proofreading Exonuclease Subunit E. coli DNA Polymerase III
To be Published
|
|
2AXD
 
 | | solution structure of the theta subunit of escherichia coli DNA polymerase III in complex with the epsilon subunit | | Descriptor: | DNA polymerase III, theta subunit | | Authors: | Keniry, M.A, Park, A.Y, Owen, E.A, Hamdan, S.M, Pintacuda, G, Otting, G, Dixon, N.E. | | Deposit date: | 2005-09-05 | | Release date: | 2006-07-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the theta subunit of Escherichia coli DNA polymerase III in complex with the epsilon subunit
J.Bacteriol., 188, 2006
|
|
5TK0
 
 | | Lysozyme structure collected with 3D printed polymer mounts | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Kilah, N.L, MacDonald, N, Bunton, G, Park, A.Y, Breadmore, M.C. | | Deposit date: | 2016-10-06 | | Release date: | 2017-04-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.795 Å) | | Cite: | 3D Printed Micrometer-Scale Polymer Mounts for Single Crystal Analysis.
Anal. Chem., 89, 2017
|
|
5MB0
 
 | | Cocktail experiment A: fragments 63, 267, and 291 in complex with Endothiapepsin | | Descriptor: | 2,5-dimethyl-N-(pyridin-4-yl)furan-3-carboxamide, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Radeva, N, Heine, A, Klebe, G. | | Deposit date: | 2016-11-07 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.149 Å) | | Cite: | Comparison of cocktail versus single soaking experimets
To Be Published
|
|
4B2T
 
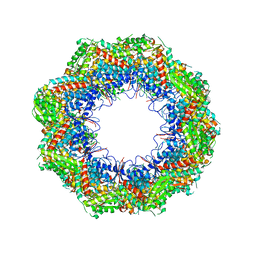 | | The crystal structures of the eukaryotic chaperonin CCT reveal its functional partitioning | | Descriptor: | T-COMPLEX PROTEIN 1 SUBUNIT ALPHA, T-COMPLEX PROTEIN 1 SUBUNIT BETA, T-COMPLEX PROTEIN 1 SUBUNIT DELTA, ... | | Authors: | Kalisman, N, Schroeder, G.F, Levitt, M. | | Deposit date: | 2012-07-17 | | Release date: | 2013-03-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | The Crystal Structures of the Eukaryotic Chaperonin Cct Reveal its Functional Partitioning
Structure, 21, 2013
|
|
2XSM
 
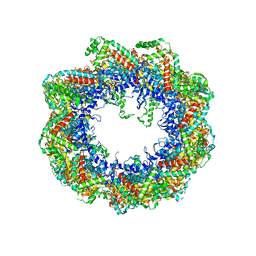 | | Crystal structure of the mammalian cytosolic chaperonin CCT in complex with tubulin | | Descriptor: | CCT | | Authors: | Munoz, I.G, Yebenes, H, Zhou, M, Mesa, P, Serna, M, Bragado-Nilsson, E, Beloso, A, Robinson, C.V, Valpuesta, J.M, Montoya, G. | | Deposit date: | 2010-10-29 | | Release date: | 2010-12-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | Crystal Structure of the Open Conformation of the Mammalian Chaperonin Cct in Complex with Tubulin.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3WZ8
 
 | | Endothiapepsin in complex with Gewald reaction-derived inhibitor (8) | | Descriptor: | DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Kuhnert, M, Steuber, H, Diederich, W.E. | | Deposit date: | 2014-09-19 | | Release date: | 2015-08-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Tracing binding modes in hit-to-lead optimization: chameleon-like poses of aspartic protease inhibitors
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
3WZ7
 
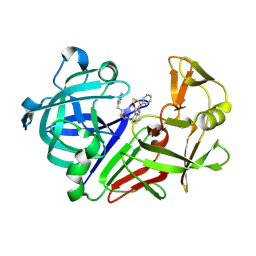 | | Endothiapepsin in complex with Gewald reaction-derived inhibitor (6) | | Descriptor: | DIMETHYL SULFOXIDE, Endothiapepsin, N-benzyl-2-({N-[2-(1H-indol-3-yl)ethyl]glycyl}amino)-4,5,6,7-tetrahydro-1-benzothiophene-3-carboxamide | | Authors: | Kuhnert, M, Steuber, H, Diederich, W.E. | | Deposit date: | 2014-09-19 | | Release date: | 2015-08-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Tracing binding modes in hit-to-lead optimization: chameleon-like poses of aspartic protease inhibitors
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
3WZ6
 
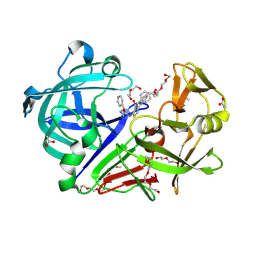 | | Endothiapepsin in complex with Gewald reaction-derived inhibitor (5) | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kuhnert, M, Steuber, H, Diederich, W.E. | | Deposit date: | 2014-09-19 | | Release date: | 2015-08-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.404 Å) | | Cite: | Tracing binding modes in hit-to-lead optimization: chameleon-like poses of aspartic protease inhibitors
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
3PSY
 
 | | Endothiapepsin in complex with an inhibitor based on the Gewald reaction | | Descriptor: | DIMETHYL SULFOXIDE, Endothiapepsin, GLYCEROL, ... | | Authors: | Koester, H, Heine, A, Klebe, G. | | Deposit date: | 2010-12-02 | | Release date: | 2011-12-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Tracing binding modes in hit-to-lead optimization: chameleon-like poses of aspartic protease inhibitors.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
3OQ9
 
 | | Structure of the FAS/FADD death domain assembly | | Descriptor: | Protein FADD, Tumor necrosis factor receptor superfamily member 6 | | Authors: | Kabaleeswaran, V, Wu, H. | | Deposit date: | 2010-09-02 | | Release date: | 2010-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (6.8 Å) | | Cite: | The Fas-FADD death domain complex structure reveals the basis of DISC assembly and disease mutations.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3RIU
 
 | |
5P27
 
 | |
5P2M
 
 | |
5P85
 
 | |
5P8K
 
 | |
5P33
 
 | |
5P3J
 
 | |
5P3X
 
 | |
5P4B
 
 | |
5P4Q
 
 | |
5P56
 
 | |
5P5L
 
 | |
5P5Z
 
 | |
5P6E
 
 | |
