5H9W
 
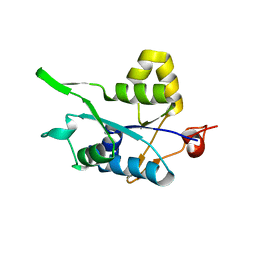 | | Crystal structure of Regnase PIN domain, form II | | Descriptor: | Ribonuclease ZC3H12A, SODIUM ION | | Authors: | Yokogawa, M, Tsushima, T, Adachi, W, Noda, N.N, Inagaki, F. | | Deposit date: | 2015-12-29 | | Release date: | 2016-03-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the regulation of enzymatic activity of Regnase-1 by domain-domain interactions
Sci Rep, 6, 2016
|
|
5H9V
 
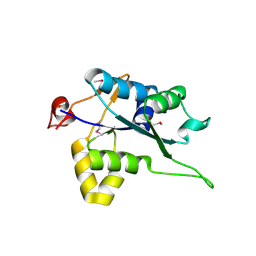 | | Crystal structure of Regnase PIN domain, form I | | Descriptor: | Ribonuclease ZC3H12A, SODIUM ION | | Authors: | Yokogawa, M, Tsushima, T, Adachi, W, Noda, N.N, Inagaki, F. | | Deposit date: | 2015-12-29 | | Release date: | 2016-03-16 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for the regulation of enzymatic activity of Regnase-1 by domain-domain interactions
Sci Rep, 6, 2016
|
|
2N5J
 
 | | Regnase-1 N-terminal domain | | Descriptor: | Ribonuclease ZC3H12A | | Authors: | Yokogawa, M, Tsushima, T, Noda, N.N, Kumeta, H, Adachi, W, Enokizono, Y, Yamashita, K, Standley, D.M, Takeuchi, O, Akira, S, Inagaki, F. | | Deposit date: | 2015-07-18 | | Release date: | 2016-03-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the regulation of enzymatic activity of Regnase-1 by domain-domain interactions
Sci Rep, 6, 2016
|
|
2N5K
 
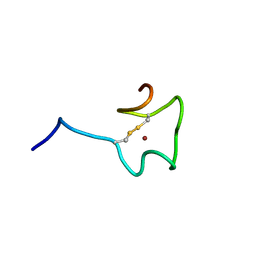 | | Regnase-1 Zinc finger domain | | Descriptor: | Ribonuclease ZC3H12A, ZINC ION | | Authors: | Yokogawa, M, Tsushima, T, Noda, N.N, Kumeta, H, Adachi, W, Enokizono, Y, Yamashita, K, Standley, D.M, Takeuchi, O, Akira, S, Inagaki, F. | | Deposit date: | 2015-07-18 | | Release date: | 2016-03-16 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the regulation of enzymatic activity of Regnase-1 by domain-domain interactions
Sci Rep, 6, 2016
|
|
2N5L
 
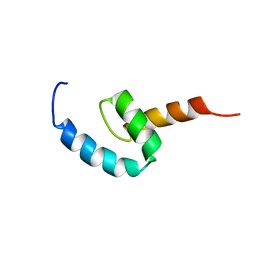 | | Regnase-1 C-terminal domain | | Descriptor: | Ribonuclease ZC3H12A | | Authors: | Yokogawa, M, Tsushima, T, Noda, N.N, Kumeta, H, Adachi, W, Enokizono, Y, Yamashita, K, Standley, D.M, Takeuchi, O, Akira, S, Inagaki, F. | | Deposit date: | 2015-07-18 | | Release date: | 2016-03-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the regulation of enzymatic activity of Regnase-1 by domain-domain interactions
Sci Rep, 6, 2016
|
|
5YGY
 
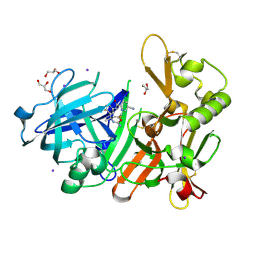 | | Crystal Structure of BACE1 in complex with (S)-N-(3-(2-amino-6-(fluoromethyl)-4 -methyl-4H-1,3-oxazin-4-yl)-4-fluorophenyl)-5-cyanopicolinamide | | Descriptor: | Beta-secretase 1, GLYCEROL, IODIDE ION, ... | | Authors: | Fuchino, K, Mitsuoka, Y, Masui, M, Kurose, N, Yoshida, S, Komano, K, Yamamoto, T, Ogawa, M, Unemura, C, Hosono, M, Ito, H, Sakaguchi, G, Ando, S, Ohnishi, S, Kido, Y, Fukushima, T, Miyajima, H, Hiroyama, S, Koyabu, K, Dhuyvetter, D, Borghys, H, Gijsen, H, Yamano, Y, Iso, Y, Kusakabe, K. | | Deposit date: | 2017-09-27 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rational Design of Novel 1,3-Oxazine Based beta-Secretase (BACE1) Inhibitors: Incorporation of a Double Bond To Reduce P-gp Efflux Leading to Robust A beta Reduction in the Brain
J. Med. Chem., 61, 2018
|
|
5YGX
 
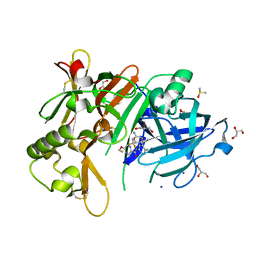 | | Structure of BACE1 in complex with N-(3-((4R,5R,6S)-2-amino-6-(1,1-difluoroethyl)-5-fluoro-4-methyl-5,6-dihydro-4H-1,3-oxazin-4-yl)-4-fluorophenyl)-5-(fluoromethoxy)pyrazine-2-carboxamide | | Descriptor: | Beta-secretase 1, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Nakahara, K, Fuchino, K, Komano, K, Asada, N, Tadano, G, Hasegawa, T, Yamamoto, T, Sako, Y, Ogawa, M, Unemura, C, Hosono, M, Sakaguchi, G, Ando, S, Ohnishi, S, Kido, Y, Fukushima, T, Dhuyvetter, D, Borghys, H, Gijsen, H, Yamano, Y, Iso, Y, Kusakabe, K. | | Deposit date: | 2017-09-27 | | Release date: | 2018-08-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of Potent and Centrally Active 6-Substituted 5-Fluoro-1,3-dihydro-oxazine beta-Secretase (BACE1) Inhibitors via Active Conformation Stabilization
J. Med. Chem., 61, 2018
|
|
7BWI
 
 | | Solution structure of recombinant APETx1 | | Descriptor: | Kappa-actitoxin-Ael2a | | Authors: | Matsumura, K, Kobayashi, N, Kurita, J, Nishimura, Y, Yokogawa, M, Imai, S, Shimada, I, Osawa, M. | | Deposit date: | 2020-04-14 | | Release date: | 2020-12-23 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Mechanism of hERG inhibition by gating-modifier toxin, APETx1, deduced by functional characterization.
Bmc Mol Cell Biol, 22, 2021
|
|
3WX0
 
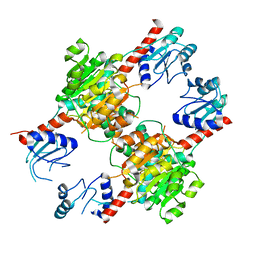 | |
1GDV
 
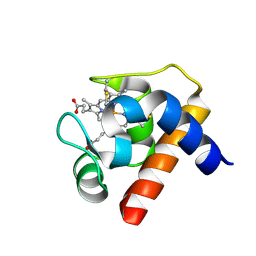 | | CRYSTAL STRUCTURE OF CYTOCHROME C6 FROM RED ALGA PORPHYRA YEZOENSIS AT 1.57 A RESOLUTION | | Descriptor: | CYTOCHROME C6, HEME C | | Authors: | Yamada, S, Park, S.-Y, Shimizu, H, Shiro, Y, Oku, T. | | Deposit date: | 2000-10-06 | | Release date: | 2001-04-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structure of cytochrome c6 from the red alga Porphyra yezoensis at 1. 57 A resolution.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
3B21
 
 | |
3VX8
 
 | |
8G0L
 
 | |
