1OBW
 
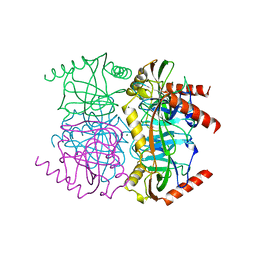 | | STRUCTURE OF INORGANIC PYROPHOSPHATASE | | Descriptor: | INORGANIC PYROPHOSPHATASE, MAGNESIUM ION | | Authors: | Oganessyan, V.Yu, Harutyunyan, E.H, Avaeva, S.M, Oganessyan, N.N, Mather, T, Huber, R. | | Deposit date: | 1996-10-09 | | Release date: | 1997-09-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of holo inorganic pyrophosphatase from Escherichia coli at 1.9 A resolution. Mechanism of hydrolysis.
Biochemistry, 36, 1997
|
|
5H9W
 
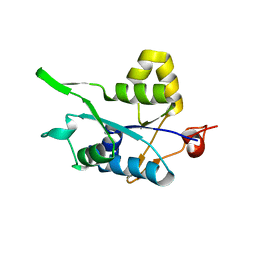 | | Crystal structure of Regnase PIN domain, form II | | Descriptor: | Ribonuclease ZC3H12A, SODIUM ION | | Authors: | Yokogawa, M, Tsushima, T, Adachi, W, Noda, N.N, Inagaki, F. | | Deposit date: | 2015-12-29 | | Release date: | 2016-03-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the regulation of enzymatic activity of Regnase-1 by domain-domain interactions
Sci Rep, 6, 2016
|
|
7ZXV
 
 | | Orange Carotenoid Protein Trp-288 BTA mutant | | Descriptor: | CHLORIDE ION, Orange carotenoid-binding protein, beta,beta-caroten-4-one, ... | | Authors: | Moldenhauer, M, Tseng, H.-W, Kraskov, A, Tavraz, N.N, Hildebrandt, P, Hochberg, G, Essen, L.-O, Budisa, N, Korf, L, Maksimov, E.G, Friedrich, T. | | Deposit date: | 2022-05-23 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Parameterization of a single H-bond in Orange Carotenoid Protein by atomic mutation reveals principles of evolutionary design of complex chemical photosystems.
Front Mol Biosci, 10, 2023
|
|
1D3V
 
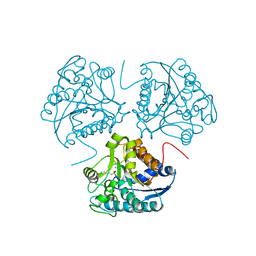 | | CRYSTAL STRUCTURE OF THE BINUCLEAR MANGANESE METALLOENZYME ARGINASE COMPLEXED WITH 2(S)-AMINO-6-BORONOHEXANOIC ACID, AN L-ARGININE ANALOG | | Descriptor: | 2(S)-AMINO-6-BORONOHEXANOIC ACID, MANGANESE (II) ION, PROTEIN (ARGINASE) | | Authors: | Cox, J.D, Kim, N.N, Traish, A.M, Christianson, D.W. | | Deposit date: | 1999-10-01 | | Release date: | 1999-11-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Arginase-boronic acid complex highlights a physiological role in erectile function.
Nat.Struct.Biol., 6, 1999
|
|
6K5S
 
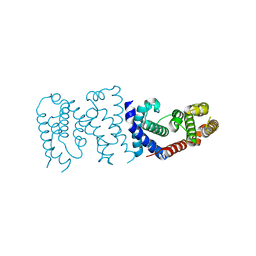 | |
7QIK
 
 | | SARS-CoV-2 Nucleocapsid phosphopeptide 193-200 bound to human 14-3-3 sigma | | Descriptor: | 14-3-3 protein sigma, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BROMIDE ION, ... | | Authors: | Sluchanko, N.N, Tugaeva, K.V, Smith, J.L.R, Antson, A.A. | | Deposit date: | 2021-12-15 | | Release date: | 2021-12-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | SARS-CoV-2 Nucleocapsid phosphopeptide 193-200 bound to human 14-3-3 sigma
To Be Published
|
|
7QIP
 
 | |
6K5U
 
 | |
4YJK
 
 | | Crystal structure of C212S mutant of Shewanella oneidensis MR-1 uridine phosphorylase | | Descriptor: | SULFATE ION, URACIL, Uridine phosphorylase | | Authors: | Safonova, T.N, Mordkovich, N.N, Manuvera, V.A, Dorovatovsky, P.V, Veiko, V.P, Popov, V.O, Polyakov, K.M. | | Deposit date: | 2015-03-03 | | Release date: | 2015-03-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Concerted action of two subunits of the functional dimer of Shewanella oneidensis MR-1 uridine phosphorylase derived from a comparison of the C212S mutant and the wild-type enzyme.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
4YK8
 
 | |
3MMP
 
 | | Structure of the Qb replicase, an RNA-dependent RNA polymerase consisting of viral and host proteins | | Descriptor: | (2S)-1-[3-{[(2R)-2-hydroxypropyl]oxy}-2,2-bis({[(2R)-2-hydroxypropyl]oxy}methyl)propoxy]propan-2-ol, Elongation factor Tu 2, Elongation factor Ts, ... | | Authors: | Kidmose, R.T, Vasiliev, N.N, Chetverin, A.B, Knudsen, C.R, Andersen, G.R. | | Deposit date: | 2010-04-20 | | Release date: | 2010-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Qbeta replicase, an RNA-dependent RNA polymerase consisting of viral and host proteins.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2Z0D
 
 | |
2Z0E
 
 | |
2JK0
 
 | | Structural and functional insights into Erwinia carotovora L- asparaginase | | Descriptor: | ASPARTIC ACID, L-ASPARAGINASE | | Authors: | Papageorgiou, A.C, Posypanova, G.A, Andersson, C.S, Sokolov, N.N, Krasotkina, J. | | Deposit date: | 2008-05-23 | | Release date: | 2008-08-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Functional Insights Into Erwinia Carotovora L-Asparaginase.
FEBS J., 275, 2008
|
|
2DYM
 
 | |
2DYT
 
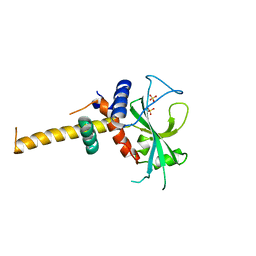 | |
2DYO
 
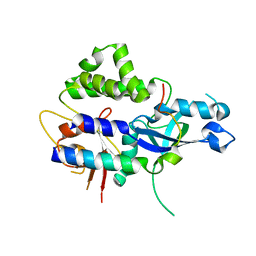 | |
2FJT
 
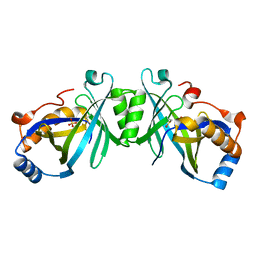 | | Adenylyl cyclase class iv from Yersinia pestis | | Descriptor: | Adenylyl cyclase class IV, SULFATE ION | | Authors: | Gallagher, D.T, Smith, N.N, Kim, S.-K, Reddy, P.T, Robinson, H, Heroux, A. | | Deposit date: | 2006-01-03 | | Release date: | 2006-11-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Structure of the class IV adenylyl cyclase reveals a novel fold
J.Mol.Biol., 362, 2006
|
|
3L2B
 
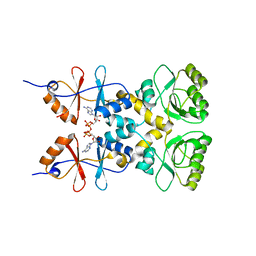 | | Crystal structure of the CBS and DRTGG domains of the regulatory region of Clostridium perfringens pyrophosphatase complexed with activator, diadenosine tetraphosphate | | Descriptor: | BIS(ADENOSINE)-5'-TETRAPHOSPHATE, Probable manganase-dependent inorganic pyrophosphatase | | Authors: | Tuominen, H, Salminen, A, Oksanen, E, Jamsen, J, Heikkila, O, Lehtio, L, Magretova, N.N, Goldman, A, Baykov, A.A, Lahti, R. | | Deposit date: | 2009-12-15 | | Release date: | 2010-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal Structures of the CBS and DRTGG Domains of the Regulatory Region of Clostridiumperfringens Pyrophosphatase Complexed with the Inhibitor, AMP, and Activator, Diadenosine Tetraphosphate.
J.Mol.Biol., 2010
|
|
3L31
 
 | | Crystal structure of the CBS and DRTGG domains of the regulatory region of Clostridium perfringens pyrophosphatase complexed with the inhibitor, AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Probable manganase-dependent inorganic pyrophosphatase | | Authors: | Tuominen, H, Salminen, A, Oksanen, E, Jamsen, J, Heikkila, O, Lehtio, L, Magretova, N.N, Goldman, A, Baykov, A.A, Lahti, R. | | Deposit date: | 2009-12-16 | | Release date: | 2010-04-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of the CBS and DRTGG Domains of the Regulatory Region of Clostridiumperfringens Pyrophosphatase Complexed with the Inhibitor, AMP, and Activator, Diadenosine Tetraphosphate.
J.Mol.Biol., 2010
|
|
6AAG
 
 | |
6AAF
 
 | |
113D
 
 | | THE STRUCTURE OF GUANOSINE-THYMIDINE MISMATCHES IN B-DNA AT 2.5 ANGSTROMS RESOLUTION | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*TP*GP*CP*G)-3') | | Authors: | Hunter, W.N, Brown, T, Kneale, G, Anand, N.N, Rabinovich, D, Kennard, O. | | Deposit date: | 1993-01-04 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of guanosine-thymidine mismatches in B-DNA at 2.5-A resolution.
J.Biol.Chem., 262, 1987
|
|
6A9E
 
 | | Crystal structure of the N-terminal domain of Atg2 | | Descriptor: | Endolysin,Autophagy-related protein 2 | | Authors: | Osawa, T, Noda, N.N. | | Deposit date: | 2018-07-13 | | Release date: | 2019-03-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.205 Å) | | Cite: | Atg2 mediates direct lipid transfer between membranes for autophagosome formation.
Nat. Struct. Mol. Biol., 26, 2019
|
|
6A9J
 
 | | Crystal structure of the PE-bound N-terminal domain of Atg2 | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Endolysin,Autophagy-related protein 2 | | Authors: | Osawa, T, Noda, N.N. | | Deposit date: | 2018-07-13 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Atg2 mediates direct lipid transfer between membranes for autophagosome formation.
Nat. Struct. Mol. Biol., 26, 2019
|
|
