1UUD
 
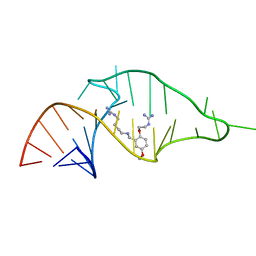 | | NMR structure of a synthetic small molecule, rbt203, bound to HIV-1 TAR RNA | | Descriptor: | N-[2-(2-{[(4-{[AMINO(IMINO)METHYL]AMINO}BUTYL)AMINO]METHYL}-4-METHOXYPHENOXY)ETHYL]GUANIDINE, RNA (5'-(*GP*GP*CP*AP*GP*AP*UP*CP*UP*GP*AP*GP *CP*CP*UP*GP*GP*GP*AP*GP*CP*UP*CP*UP*CP*UP*GP*CP*C) -3') | | Authors: | Davis, B, Afshar, M, Varani, G, Karn, J, Murchie, A.I.H, Lentzen, G, Drysdale, M.J, Potter, A.J, Bower, J, Aboul-Ela, F. | | Deposit date: | 2003-12-18 | | Release date: | 2004-03-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Rational Design of Inhibitors of HIV-1 Tar RNA Through the Stabilisation of Electrostatic "Hot Spots"
J.Mol.Biol., 336, 2004
|
|
1OLN
 
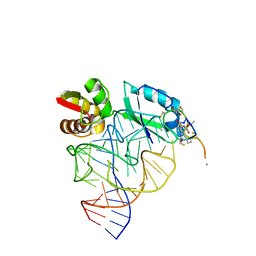 | | Model for thiostrepton antibiotic binding to L11 substrate from 50S ribosomal RNA | | Descriptor: | 50S RIBOSOMAL PROTEIN L11, RNA, THIOSTREPTON | | Authors: | Lentzen, G, Klinck, R, Matassova, N, Aboul-Ela, F, Murchie, A.I.H. | | Deposit date: | 2003-08-08 | | Release date: | 2003-09-11 | | Last modified: | 2019-08-21 | | Method: | SOLUTION NMR, THEORETICAL MODEL | | Cite: | Structural Basis for Contrasting Activities of Ribosome Binding Thiazole Antibiotics
Chem.Biol., 10, 2003
|
|
1UUI
 
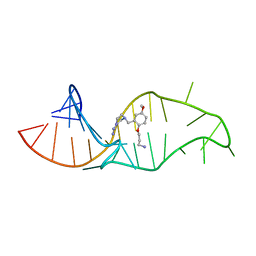 | | NMR structure of a synthetic small molecule, rbt158, bound to HIV-1 TAR RNA | | Descriptor: | 4-[AMINO(IMINO)METHYL]-1-[2-(3-AMMONIOPROPOXY)-5-METHOXYBENZYL]PIPERAZIN-1-IUM, 5'-R(*GP*GP*CP*AP*GP*AP*UP*CP*UP*GP*AP*GP*CP* CP*UP*GP*GP*GP*AP*GP*CP*UP*CP*UP*CP*UP*GP*CP*C)-3' | | Authors: | Davis, B, Afshar, M, Varani, G, Karn, J, Murchie, A.I.H, Lentzen, G, Drysdale, M.J, Potter, A.J, Bower, J, Aboul-Ela, F. | | Deposit date: | 2003-12-19 | | Release date: | 2004-02-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Rational Design of Inhibitors of HIV-1 Tar RNA Through the Stabilisation of Electrostatic "Hot Spots"
J.Mol.Biol., 336, 2004
|
|
7DLZ
 
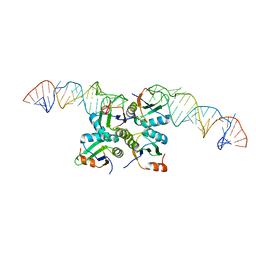 | | Crystal Structure of Methyltransferase Ribozyme | | Descriptor: | RNA (45-MER), U1 small nuclear ribonucleoprotein A | | Authors: | Gan, J.H, Gao, Y.Q, Jiang, H.Y, Chen, D.R, Murchie, A.I.H. | | Deposit date: | 2020-11-30 | | Release date: | 2021-10-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | The identification and characterization of a selected SAM-dependent methyltransferase ribozyme that is present in natural sequences
Nat Catal, 4, 2021
|
|
352D
 
 | | THE CRYSTAL STRUCTURE OF A PARALLEL-STRANDED PARALLEL-STRANDED GUANINE TETRAPLEX AT 0.95 ANGSTROM RESOLUTION | | Descriptor: | CALCIUM ION, DNA (5'-D(*TP*GP*GP*GP*GP*T)-3'), SODIUM ION | | Authors: | Phillips, K, Dauter, Z, Murchie, A.I.H, Lilley, D.M.J, Luisi, B. | | Deposit date: | 1997-09-04 | | Release date: | 1997-11-10 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | The crystal structure of a parallel-stranded guanine tetraplex at 0.95 A resolution.
J.Mol.Biol., 273, 1997
|
|
244D
 
 | | THE HIGH-RESOLUTION CRYSTAL STRUCTURE OF A PARALLEL-STRANDED GUANINE TETRAPLEX | | Descriptor: | CALCIUM ION, DNA (5'-D(*TP*GP*GP*GP*GP*T)-3'), SODIUM ION | | Authors: | Laughlan, G, Murchie, A.I.H, Norman, D.G, Moore, M.H, Moody, P.C.E, Lilley, D.M.J, Luisi, B. | | Deposit date: | 1995-10-19 | | Release date: | 1996-02-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The high-resolution crystal structure of a parallel-stranded guanine tetraplex.
Science, 265, 1994
|
|
7DWH
 
 | | Complex structure of SAM-dependent methyltransferase ribozyme | | Descriptor: | COPPER (II) ION, RNA (45-MER), S-ADENOSYLMETHIONINE, ... | | Authors: | Jiang, H.Y, Gao, Y.Q, Chen, D.R, Murchie, A. | | Deposit date: | 2021-01-17 | | Release date: | 2021-10-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The identification and characterization of a selected SAM-dependent methyltransferase ribozyme that is present in natural sequences
Nat Catal, 4, 2021
|
|
3NK7
 
 | | Structure of the Nosiheptide-resistance methyltransferase S-adenosyl-L-methionine Complex | | Descriptor: | 23S rRNA methyltransferase, S-ADENOSYLMETHIONINE | | Authors: | Yang, H, Wang, Z, Shen, Y, Wang, P, Murchie, A, Xu, Y. | | Deposit date: | 2010-06-18 | | Release date: | 2010-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Nosiheptide-Resistance Methyltransferase of Streptomyces actuosus
Biochemistry, 49, 2010
|
|
3NK6
 
 | | Structure of the Nosiheptide-resistance methyltransferase | | Descriptor: | 23S rRNA methyltransferase | | Authors: | Yang, H, Wang, Z, Shen, Y, Wang, P, Murchie, A, Xu, Y. | | Deposit date: | 2010-06-18 | | Release date: | 2010-07-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Nosiheptide-Resistance Methyltransferase of Streptomyces actuosus
Biochemistry, 49, 2010
|
|
1UTS
 
 | | Designed HIV-1 TAR Binding Ligand | | Descriptor: | N-[2-(3-AMINOPROPOXY)-5-(1H-INDOL-5-YL)BENZYL]-N-(2-PIPERAZIN-1-YLETHYL)AMINE, RNA (5'-(*GP*GP*CP*AP*GP*AP*UP*CP*UP*GP*AP*GP *CP*CP*UP*GP*GP*GP*AP*GP*CP*UP*CP*UP*CP*UP*GP*CP*C) -3') | | Authors: | Davis, B, Murchie, A.I.H, Aboul-Ela, F, Karn, J. | | Deposit date: | 2003-12-10 | | Release date: | 2004-02-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure-based drug design targeting an inactive RNA conformation: exploiting the flexibility of HIV-1 TAR RNA.
J.Mol.Biol., 336, 2004
|
|
