3TTQ
 
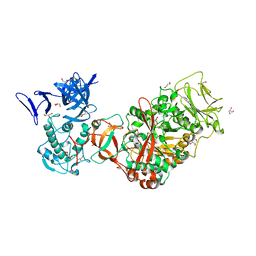 | | Crystal structure of Leuconostoc mesenteroides NRRL B-1299 N-terminally truncated dextransucrase DSR-E in orthorhombic apo-form at 1.9 angstrom resolution | | Descriptor: | CALCIUM ION, Dextransucrase, GLYCEROL, ... | | Authors: | Brison, Y, Pijning, T, Fabre, E, Mourey, L, Morel, S, Potocki-Veronese, G, Monsan, P, Remaud-Simeon, M, Dijkstra, B.W, Tranier, S. | | Deposit date: | 2011-09-15 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Functional and structural characterization of alpha-(1-2) branching sucrase derived from DSR-E glucansucrase
J.Biol.Chem., 287, 2012
|
|
3TTO
 
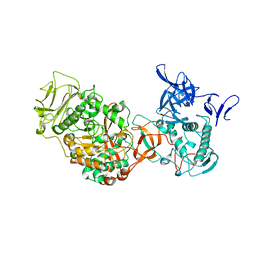 | | Crystal structure of Leuconostoc mesenteroides NRRL B-1299 N-terminally truncated dextransucrase DSR-E in triclinic form | | Descriptor: | CALCIUM ION, Dextransucrase, GLYCEROL | | Authors: | Brison, Y, Pijning, T, Fabre, E, Mourey, L, Morel, S, Potocki-Veronese, G, Monsan, P, Tranier, S, Remaud-Simeon, M, Dijkstra, B.W. | | Deposit date: | 2011-09-15 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Functional and structural characterization of alpha-(1-2) branching sucrase derived from DSR-E glucansucrase
J.Biol.Chem., 287, 2012
|
|
4FLQ
 
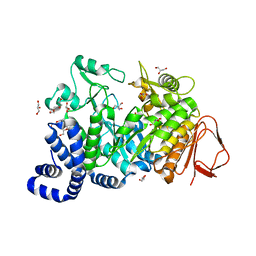 | | Crystal structure of Amylosucrase double mutant A289P-F290I from Neisseria polysaccharea. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Amylosucrase, GLYCEROL, ... | | Authors: | Guerin, F, Champion, E, Moulis, C, Barbe, S, Tran, T.H, Morel, S, Descroix, K, Monsan, P, Mulard, L.A, Remaud-Simeon, M, Andre, I, Mourey, L, Tranier, S. | | Deposit date: | 2012-06-15 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Applying pairwise combinations of amino Acid mutations for sorting out highly efficient glucosylation tools for chemo-enzymatic synthesis of bacterial oligosaccharides.
J.Am.Chem.Soc., 134, 2012
|
|
4FLO
 
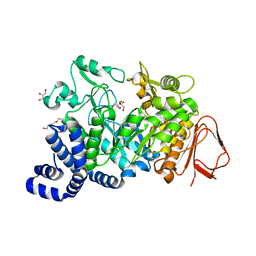 | | Crystal structure of Amylosucrase double mutant A289P-F290C from Neisseria polysaccharea | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Amylosucrase, GLYCEROL, ... | | Authors: | Guerin, F, Champion, E, Moulis, C, Barbe, S, Tran, T.H, Morel, S, Descroix, K, Monsan, P, Mulard, L.A, Remaud-Simeon, M, Andre, I, Mourey, L, Tranier, S. | | Deposit date: | 2012-06-15 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Applying pairwise combinations of amino Acid mutations for sorting out highly efficient glucosylation tools for chemo-enzymatic synthesis of bacterial oligosaccharides.
J.Am.Chem.Soc., 134, 2012
|
|
4FLS
 
 | | Crystal structure of Amylosucrase inactive double mutant F290K-E328Q from Neisseria polysaccharea in complex with sucrose. | | Descriptor: | Amylosucrase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Guerin, F, Champion, E, Moulis, C, Barbe, S, Tran, T.H, Morel, S, Descroix, K, Monsan, P, Mulard, L.A, Remaud-Simeon, M, Andre, I, Mourey, L, Tranier, S. | | Deposit date: | 2012-06-15 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Applying pairwise combinations of amino Acid mutations for sorting out highly efficient glucosylation tools for chemo-enzymatic synthesis of bacterial oligosaccharides.
J.Am.Chem.Soc., 134, 2012
|
|
4FLR
 
 | | Crystal structure of Amylosucrase double mutant A289P-F290L from Neisseria polysaccharea | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Amylosucrase, GLYCEROL, ... | | Authors: | Guerin, F, Champion, E, Moulis, C, Barbe, S, Tran, T.H, Morel, S, Descroix, K, Monsan, P, Mulard, L.A, Remaud-Simeon, M, Andre, I, Mourey, L, Tranier, S. | | Deposit date: | 2012-06-15 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Applying pairwise combinations of amino Acid mutations for sorting out highly efficient glucosylation tools for chemo-enzymatic synthesis of bacterial oligosaccharides.
J.Am.Chem.Soc., 134, 2012
|
|
4UDJ
 
 | | Crystal structure of b-1,4-mannopyranosyl-chitobiose phosphorylase at 1.60 Angstrom in complex with beta-D-mannopyranose and inorganic phosphate | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Ladeveze, S, Cioci, G, Potocki-Veronese, G, Tranier, S, Mourey, L. | | Deposit date: | 2014-12-10 | | Release date: | 2015-05-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural Bases for N-Glycan Processing by Mannoside Phosphorylase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4UDK
 
 | | Crystal structure of b-1,4-mannopyranosyl-chitobiose phosphorylase at 1.76 Angstrom from unknown human gut bacteria (Uhgb_MP) in complex with N-acetyl-D-glucosamine, beta-D-mannopyranose and inorganic phosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-glucopyranose, GLYCEROL, ... | | Authors: | Ladeveze, S, Cioci, G, Potocki-Veronese, G, Tranier, S, Mourey, L. | | Deposit date: | 2014-12-10 | | Release date: | 2015-05-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural Bases for N-Glycan Processing by Mannoside Phosphorylase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4UDI
 
 | | Crystal structure of b-1,4-mannopyranosyl-chitobiose phosphorylase at 1.85 Angstrom from unknown human gut bacteria (Uhgb_MP) | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Ladeveze, S, Cioci, G, Potocki-Veronese, G, Tranier, S, Mourey, L. | | Deposit date: | 2014-12-10 | | Release date: | 2015-05-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Bases for N-Glycan Processing by Mannoside Phosphorylase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6HVG
 
 | |
6SZI
 
 | | ASR Alternansucrase in complex with isomaltose | | Descriptor: | Alternansucrase, CALCIUM ION, alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose | | Authors: | Cioci, G, Molina, M, Moulis, C, Remaud-Simeon, M. | | Deposit date: | 2019-10-02 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A specific oligosaccharide-binding site in the alternansucrase catalytic domain mediates alternan elongation.
J.Biol.Chem., 295, 2020
|
|
6T16
 
 | | ASR Alternansucrase in complex with panose | | Descriptor: | Alternansucrase, CALCIUM ION, alpha-D-glucopyranose, ... | | Authors: | Cioci, G, Molina, M, Moulis, C, Remaud-Simeon, M. | | Deposit date: | 2019-10-03 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A specific oligosaccharide-binding site in the alternansucrase catalytic domain mediates alternan elongation.
J.Biol.Chem., 295, 2020
|
|
6SYQ
 
 | | ASR Alternansucrase in complex with isomaltotriose | | Descriptor: | Alternansucrase, CALCIUM ION, alpha-D-glucopyranose, ... | | Authors: | Cioci, G, Molina, M, Moulis, C, Remaud-Simeon, M. | | Deposit date: | 2019-09-30 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A specific oligosaccharide-binding site in the alternansucrase catalytic domain mediates alternan elongation.
J.Biol.Chem., 295, 2020
|
|
6T18
 
 | | ASR Alternansucrase in complex with oligoalternan | | Descriptor: | Alternansucrase, CALCIUM ION, alpha-D-glucopyranose-(1-3)-alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose, ... | | Authors: | Cioci, G, Molina, M, Moulis, C, Remaud-Simeon, M. | | Deposit date: | 2019-10-03 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | A specific oligosaccharide-binding site in the alternansucrase catalytic domain mediates alternan elongation.
J.Biol.Chem., 295, 2020
|
|
6T1P
 
 | | ASR Alternansucrase in complex with isomaltononaose | | Descriptor: | Alternansucrase, CALCIUM ION, alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose, ... | | Authors: | Cioci, G, Molina, M, Moulis, C, Remaud-Simeon, M. | | Deposit date: | 2019-10-04 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | A specific oligosaccharide-binding site in the alternansucrase catalytic domain mediates alternan elongation.
J.Biol.Chem., 295, 2020
|
|
