5JVJ
 
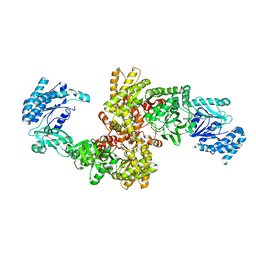 | | C4-type pyruvate phosphate dikinase: different conformational states of the nucleotide binding domain in the dimer | | Descriptor: | MAGNESIUM ION, PHOSPHOENOLPYRUVATE, Pyruvate, ... | | Authors: | Minges, A, Hoeppner, A, Groth, G. | | Deposit date: | 2016-05-11 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.898 Å) | | Cite: | Structural intermediates and directionality of the swiveling motion of Pyruvate Phosphate Dikinase.
Sci Rep, 7, 2017
|
|
5JVN
 
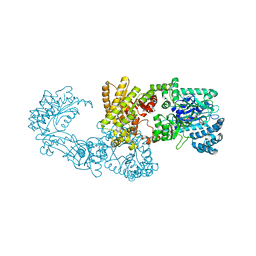 | | C3-type pyruvate phosphate dikinase: intermediate state of the central domain in the swiveling mechanism | | Descriptor: | 2'-Bromo-2'-deoxyadenosine 5'-[beta,gamma-imide]triphosphoric acid, MAGNESIUM ION, PHOSPHOENOLPYRUVATE, ... | | Authors: | Minges, A, Hoeppner, A, Groth, G. | | Deposit date: | 2016-05-11 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural intermediates and directionality of the swiveling motion of Pyruvate Phosphate Dikinase.
Sci Rep, 7, 2017
|
|
5JVL
 
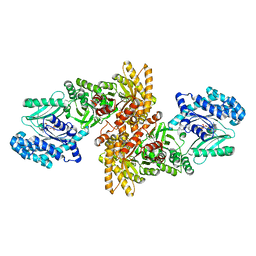 | | C4-type pyruvate phospate dikinase: nucleotide binding domain with bound ATP analogue | | Descriptor: | 2'-Bromo-2'-deoxyadenosine 5'-[beta,gamma-imide]triphosphoric acid, MAGNESIUM ION, PHOSPHOENOLPYRUVATE, ... | | Authors: | Minges, A, Hoeppner, A, Groth, G. | | Deposit date: | 2016-05-11 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural intermediates and directionality of the swiveling motion of Pyruvate Phosphate Dikinase.
Sci Rep, 7, 2017
|
|
5LU4
 
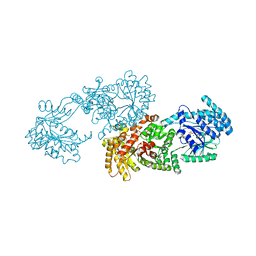 | | C4-type pyruvate phosphate dikinase: conformational intermediate of central domain in the swiveling mechanism | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PYRUVIC ACID, ... | | Authors: | Minges, A, Hoeppner, A, Groth, G. | | Deposit date: | 2016-09-08 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Trapped intermediate state of plant pyruvate phosphate dikinase indicates substeps in catalytic swiveling domain mechanism.
Protein Sci., 26, 2017
|
|
4ZDV
 
 | |
7F80
 
 | |
3WMD
 
 | | Crystal structure of epoxide hydrolase MonBI | | Descriptor: | Probable monensin biosynthesis isomerase | | Authors: | Minami, A, Ose, T, Sato, K, Oikawa, A, Kuroki, K, Maenaka, K, Oguri, H, Oikawa, H. | | Deposit date: | 2013-11-16 | | Release date: | 2014-01-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Allosteric regulation of epoxide opening cascades by a pair of epoxide hydrolases in monensin biosynthesis
Acs Chem.Biol., 9, 2014
|
|
7POG
 
 | | High-resolution structure of native toxin A from Clostridioides difficile | | Descriptor: | Toxin A, ZINC ION | | Authors: | Boesen, T, Joergensen, R, Aminzadeh, A, Engelbrecht Larsen, C. | | Deposit date: | 2021-09-08 | | Release date: | 2021-12-08 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | High-resolution structure of native toxin A from Clostridioides difficile.
Embo Rep., 23, 2022
|
|
7BBM
 
 | | Mutant nitrobindin M75L/H76L/Q96C/M148L (NB4H) from Arabidopsis thaliana with cofactor MnPPIX | | Descriptor: | 1,2-ETHANEDIOL, MANGANESE PROTOPORPHYRIN IX, UPF0678 fatty acid-binding protein-like protein At1g79260 | | Authors: | Minges, A, Sauer, D.F, Wittwer, M, Markel, U, Spiertz, M, Schiffels, J, Davari, M.D, Okuda, J, Schwaneberg, U, Groth, G. | | Deposit date: | 2020-12-18 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Chemogenetic engineering of nitrobindin toward an artificial epoxygenase
Catalysis Science And Technology, 2021
|
|
3AMO
 
 | | Time-resolved X-ray Crystal Structure Analysis of Enzymatic Reaction of Copper Amine Oxidase from Arthrobacter globiformis | | Descriptor: | COPPER (II) ION, GLYCEROL, Phenylethylamine oxidase, ... | | Authors: | Kataoka, M, Oya, H, Tominaga, A, Otsu, M, Okajima, T, Tanizawa, K, Yamaguchi, H. | | Deposit date: | 2010-08-20 | | Release date: | 2011-11-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Detection of the reaction intermediates catalyzed by a copper amine oxidase.
J.SYNCHROTRON RADIAT., 18, 2011
|
|
3COD
 
 | | Crystal Structure of T90A/D115A mutant of Bacteriorhodopsin | | Descriptor: | Bacteriorhodopsin, RETINAL | | Authors: | Joh, N.H, Min, A, Faham, S, Bowie, J.U. | | Deposit date: | 2008-03-27 | | Release date: | 2008-04-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Modest stabilization by most hydrogen-bonded side-chain interactions in membrane proteins.
Nature, 453, 2008
|
|
8WDV
 
 | | Photosynthetic LH1-RC complex from the purple sulfur bacterium Allochromatium vinosum purified by Ca2+-DEAE | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (2S)-3-hydroxypropane-1,2-diyl dihexadecanoate, Antenna complex alpha/beta subunit, ... | | Authors: | Tani, K, Kanno, R, Harada, A, Kobayashi, A, Minamino, A, Nakamura, N, Ji, X.-C, Purba, E.R, Hall, M, Yu, L.-J, Madigan, M.T, Mizoguchi, A, Iwasaki, K, Humbel, B.M, Kimura, Y, Wang-Otomo, Z.-Y. | | Deposit date: | 2023-09-16 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.24 Å) | | Cite: | High-resolution structure and biochemical properties of the LH1-RC photocomplex from the model purple sulfur bacterium, Allochromatium vinosum.
Commun Biol, 7, 2024
|
|
8WDU
 
 | | Photosynthetic LH1-RC complex from the purple sulfur bacterium Allochromatium vinosum purified by sucrose density | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (2S)-3-hydroxypropane-1,2-diyl dihexadecanoate, Antenna complex alpha/beta subunit, ... | | Authors: | Tani, K, Kanno, R, Harada, A, Kobayashi, A, Minamino, A, Nakamura, N, Ji, X.-C, Purba, E.R, Hall, M, Yu, L.-J, Madigan, M.T, Mizoguchi, A, Iwasaki, K, Humbel, B.M, Kimura, Y, Wang-Otomo, Z.-Y. | | Deposit date: | 2023-09-16 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.24 Å) | | Cite: | High-resolution structure and biochemical properties of the LH1-RC photocomplex from the model purple sulfur bacterium, Allochromatium vinosum.
Commun Biol, 7, 2024
|
|
6AHE
 
 | | Crystal structure of enoyl-ACP reductase from Acinetobacter baumannii in complex with NAD and AFN-1252 | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADH], N-methyl-N-[(3-methyl-1-benzofuran-2-yl)methyl]-3-(7-oxo-5,6,7,8-tetrahydro-1,8-naphthyridin-3-yl)propanamide, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Rani, S.T, Nataraj, V, Laxminarasimhan, A, Thomas, A, Krishnamurthy, N. | | Deposit date: | 2018-08-17 | | Release date: | 2019-08-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Ternary complex formation of AFN-1252 with Acinetobacter baumannii FabI and NADH: Crystallographic and biochemical studies.
Chem.Biol.Drug Des., 96, 2020
|
|
6AH9
 
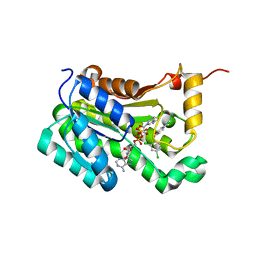 | | Crystal structure of enoyl-ACP reductase from Acinetobacter baumannii in complex with NAD and Triclosan | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRICLOSAN | | Authors: | Rani, S.T, Nataraj, V, Laxminarasimhan, A, Thomas, A, Krishnamurthy, N. | | Deposit date: | 2018-08-17 | | Release date: | 2019-08-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structure of enoyl-ACP reductase from Acinetobacter baumannii in complex with NAD and Triclosan
To Be Published
|
|
3L2B
 
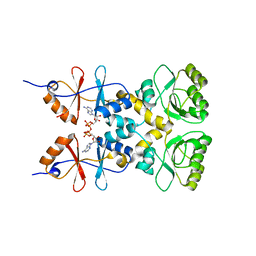 | | Crystal structure of the CBS and DRTGG domains of the regulatory region of Clostridium perfringens pyrophosphatase complexed with activator, diadenosine tetraphosphate | | Descriptor: | BIS(ADENOSINE)-5'-TETRAPHOSPHATE, Probable manganase-dependent inorganic pyrophosphatase | | Authors: | Tuominen, H, Salminen, A, Oksanen, E, Jamsen, J, Heikkila, O, Lehtio, L, Magretova, N.N, Goldman, A, Baykov, A.A, Lahti, R. | | Deposit date: | 2009-12-15 | | Release date: | 2010-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal Structures of the CBS and DRTGG Domains of the Regulatory Region of Clostridiumperfringens Pyrophosphatase Complexed with the Inhibitor, AMP, and Activator, Diadenosine Tetraphosphate.
J.Mol.Biol., 2010
|
|
3L31
 
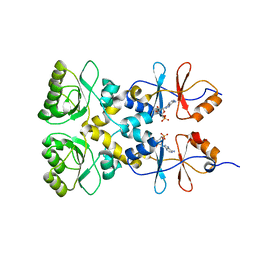 | | Crystal structure of the CBS and DRTGG domains of the regulatory region of Clostridium perfringens pyrophosphatase complexed with the inhibitor, AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Probable manganase-dependent inorganic pyrophosphatase | | Authors: | Tuominen, H, Salminen, A, Oksanen, E, Jamsen, J, Heikkila, O, Lehtio, L, Magretova, N.N, Goldman, A, Baykov, A.A, Lahti, R. | | Deposit date: | 2009-12-16 | | Release date: | 2010-04-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of the CBS and DRTGG Domains of the Regulatory Region of Clostridiumperfringens Pyrophosphatase Complexed with the Inhibitor, AMP, and Activator, Diadenosine Tetraphosphate.
J.Mol.Biol., 2010
|
|
6U9I
 
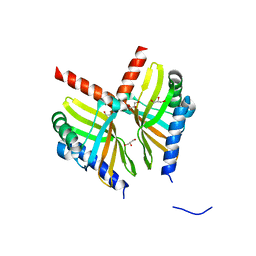 | | Crystal structure of BvnE pinacolase from Penicillium brevicompactum | | Descriptor: | BvnE, DI(HYDROXYETHYL)ETHER, GLYCEROL | | Authors: | Ye, Y, Du, L, Zhang, X, Newmister, S.A, McCauley, M, Alegre-Requena, J.V, Zhang, W, Mu, S, Minami, A, Fraley, A.E, Adrover-Castellano, M.L, Carney, N, Shende, V.V, Oikawa, H, Kato, H, Tsukamoto, S, Paton, R.S, Williams, R.M, Sherman, D.H, Li, S. | | Deposit date: | 2019-09-09 | | Release date: | 2020-09-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.777 Å) | | Cite: | Fungal-derived brevianamide assembly by a stereoselective semipinacolase.
Nat Catal, 3, 2020
|
|
4JOA
 
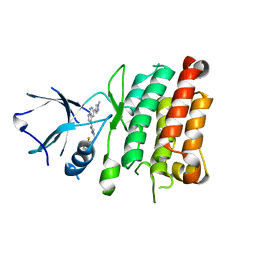 | | Crystal Structure of Human Anaplastic Lymphoma Kinase in complex with 7-azaindole based inhibitor | | Descriptor: | 3-[1-(2,5-difluorobenzyl)-1H-pyrazol-4-yl]-5-(1-methyl-1H-pyrazol-4-yl)-1H-pyrrolo[2,3-b]pyridine, ALK tyrosine kinase receptor | | Authors: | Hosahalli, S, Krishnamurthy, N.R, Lakshminarasimhan, A. | | Deposit date: | 2013-03-18 | | Release date: | 2013-07-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of 7-azaindole based anaplastic lymphoma kinase (ALK) inhibitors: wild type and mutant (L1196M) active compounds with unique binding mode
Bioorg.Med.Chem.Lett., 23, 2013
|
|
5Y79
 
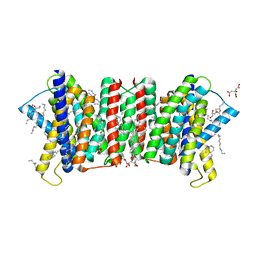 | | Crystal structure of the triose-phosphate/phosphate translocator in complex with 3-phosphoglycerate | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3-PHOSPHOGLYCERIC ACID, CITRATE ANION, ... | | Authors: | Lee, Y, Nishizawa, T, Takemoto, M, Kumazaki, K, Yamashita, K, Hirata, K, Minoda, A, Nagatoishi, S, Tsumoto, K, Ishitani, R, Nureki, O. | | Deposit date: | 2017-08-16 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the triose-phosphate/phosphate translocator reveals the basis of substrate specificity
Nat Plants, 3, 2017
|
|
4F86
 
 | | Structure analysis of Geranyl diphosphate methyltransferase in complex with GPP and sinefungin | | Descriptor: | GERANYL DIPHOSPHATE, Geranyl diphosphate 2-C-methyltransferase, MAGNESIUM ION, ... | | Authors: | Ariyawutthiphan, O, Ose, T, Minami, A, Gao, Y.G, Yao, M, Oikawa, H, Tanaka, I. | | Deposit date: | 2012-05-17 | | Release date: | 2012-10-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure analysis of geranyl pyrophosphate methyltransferase and the proposed reaction mechanism of SAM-dependent C-methylation
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4F85
 
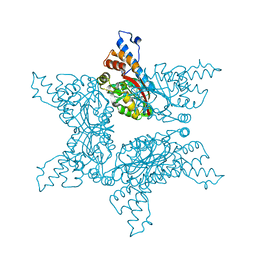 | | Structure analysis of Geranyl diphosphate methyltransferase | | Descriptor: | Geranyl diphosphate 2-C-methyltransferase | | Authors: | Ariyawutthiphan, O, Ose, T, Minami, A, Gao, Y.G, Yao, M, Oikawa, H, Tanaka, I. | | Deposit date: | 2012-05-17 | | Release date: | 2012-10-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure analysis of geranyl pyrophosphate methyltransferase and the proposed reaction mechanism of SAM-dependent C-methylation
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4F84
 
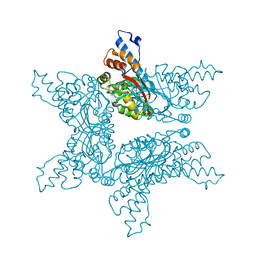 | | Structure analysis of Geranyl diphosphate methyltransferase in complex with SAM | | Descriptor: | Geranyl diphosphate 2-C-methyltransferase, S-ADENOSYLMETHIONINE | | Authors: | Ariyawutthiphan, O, Ose, T, Minami, A, Gao, Y.G, Yao, M, Oikawa, H, Tanaka, I. | | Deposit date: | 2012-05-17 | | Release date: | 2012-10-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure analysis of geranyl pyrophosphate methyltransferase and the proposed reaction mechanism of SAM-dependent C-methylation
Acta Crystallogr.,Sect.D, 68, 2012
|
|
5Y78
 
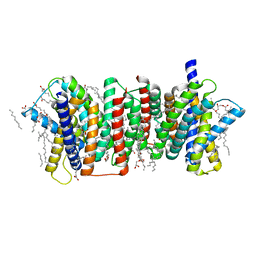 | | Crystal structure of the triose-phosphate/phosphate translocator in complex with inorganic phosphate | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, PHOSPHATE ION, Putative hexose phosphate translocator | | Authors: | Lee, Y, Nishizawa, T, Takemoto, M, Kumazaki, K, Yamashita, K, Hirata, K, Minoda, A, Nagatoishi, S, Tsumoto, K, Ishitani, R, Nureki, O. | | Deposit date: | 2017-08-16 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the triose-phosphate/phosphate translocator reveals the basis of substrate specificity
Nat Plants, 3, 2017
|
|
1WPP
 
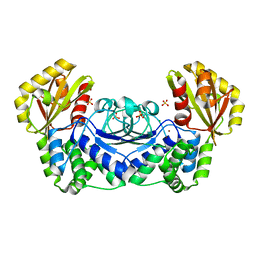 | | Structure of Streptococcus gordonii inorganic pyrophosphatase | | Descriptor: | CHLORIDE ION, Probable manganese-dependent inorganic pyrophosphatase, SULFATE ION, ... | | Authors: | Fabrichniy, I.P, Lehtio, L, Salminen, A, Baykov, A.A, Lahti, R, Goldman, A. | | Deposit date: | 2004-09-09 | | Release date: | 2004-11-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Studies of Metal Ions in Family II Pyrophosphatases: The Requirement for a Janus Ion
Biochemistry, 43, 2004
|
|
