6Z20
 
 | | Structure of the EC2 domain of CD9 in complex with nanobody 4C8 | | Descriptor: | CD9 antigen, CHLORIDE ION, GLYCEROL, ... | | Authors: | Oosterheert, W, Manshande, J, Pearce, N.M, Lutz, M, Gros, P. | | Deposit date: | 2020-05-14 | | Release date: | 2020-09-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Implications for tetraspanin-enriched microdomain assembly based on structures of CD9 with EWI-F.
Life Sci Alliance, 3, 2020
|
|
7QBA
 
 | | CryoEM structure of the ABC transporter NosDFY complexed with nitrous oxide reductase NosZ | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Zipfel, S, Mueller, C, Topitsch, A, Lutz, M, Zhang, L, Einsle, O. | | Deposit date: | 2021-11-18 | | Release date: | 2022-08-03 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Molecular interplay of an assembly machinery for nitrous oxide reductase.
Nature, 608, 2022
|
|
7ANY
 
 | | Structure of the Laspartomycin C Friulimicin-like mutant in complex with Geranyl phosphate | | Descriptor: | CADMIUM ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Zeronian, M.R, Pearce, N.M, Lutz, M, Wood, T.M, Martin, N.I, Janssen, B.J.C. | | Deposit date: | 2020-10-13 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.135 Å) | | Cite: | Mechanistic insights into the C55-P targeting lipopeptide antibiotics revealed by structure-activity studies and high-resolution crystal structures
Chem Sci, 13, 2022
|
|
6RLR
 
 | | Crystal structure of CD9 large extracellular loop | | Descriptor: | CD9 antigen | | Authors: | Neviani, V, Kroon-Batenburg, L, Lutz, M, Pearce, N.M, Pos, W, Schotte, R, Spits, H, Gros, P. | | Deposit date: | 2019-05-02 | | Release date: | 2020-09-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Implications for tetraspanin-enriched microdomain assembly based on structures of CD9 with EWI-F.
Life Sci Alliance, 3, 2020
|
|
3EF3
 
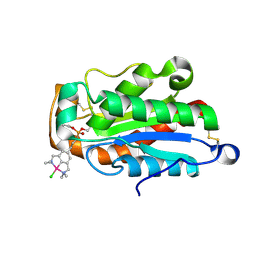 | | cut-1a; NCN-Pt-Pincer-Cutinase Hybrid | | Descriptor: | (2,6-bis[(dimethylamino-kappaN)methyl]-4-{3-[(S)-ethoxy(4-nitrophenoxy)phosphoryl]propyl}phenyl-kappaC~1~)(chloro)platinum(2+), Cutinase-1 | | Authors: | Rutten, L, Mannie, J.P.B.A, Lutz, M, Gros, P. | | Deposit date: | 2008-09-08 | | Release date: | 2009-07-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Solid-state structural characterization of cutinase-ECE-pincer-metal hybrids
Chemistry, 15, 2009
|
|
3ESD
 
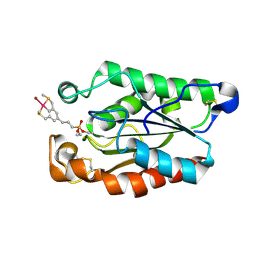 | | cut-2b; NCN-Pt-Pincer-Cutinase Hybrid | | Descriptor: | Cutinase 1, bromo(4-{3-[(R)-ethoxy(4-nitrophenoxy)phosphoryl]propyl}-2,6-bis[(methylsulfanyl-kappaS)methyl]phenyl-kappaC~1~)palladium(2+) | | Authors: | Rutten, L, Mannie, J.P.B.A, Lutz, M, Gros, P. | | Deposit date: | 2008-10-05 | | Release date: | 2009-07-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Solid-state structural characterization of cutinase-ECE-pincer-metal hybrids
Chemistry, 15, 2009
|
|
3ESC
 
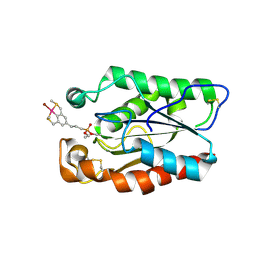 | | cut-2a; NCN-Pt-Pincer-Cutinase Hybrid | | Descriptor: | Cutinase 1, bromo(4-{3-[(R)-ethoxy(4-nitrophenoxy)phosphoryl]propyl}-2,6-bis[(methylsulfanyl-kappaS)methyl]phenyl-kappaC~1~)palladium(2+) | | Authors: | Rutten, L, Mannie, J.P.B.A, Lutz, M, Gros, P. | | Deposit date: | 2008-10-05 | | Release date: | 2009-07-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Solid-state structural characterization of cutinase-ECE-pincer-metal hybrids
Chemistry, 15, 2009
|
|
3ESA
 
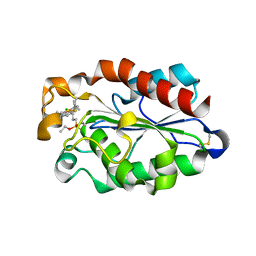 | | cut-1b; NCN-Pt-Pincer-Cutinase Hybrid | | Descriptor: | (2,6-bis[(dimethylamino-kappaN)methyl]-4-{3-[(S)-ethoxy(4-nitrophenoxy)phosphoryl]propyl}phenyl-kappaC~1~)(chloro)platinum(2+), Cutinase 1 | | Authors: | Rutten, L, Mannie, J.P.B.A, Lutz, M, Gros, P. | | Deposit date: | 2008-10-05 | | Release date: | 2009-07-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Solid-state structural characterization of cutinase-ECE-pincer-metal hybrids
Chemistry, 15, 2009
|
|
3ESB
 
 | | cut-1c; NCN-Pt-Pincer-Cutinase Hybrid | | Descriptor: | (2,6-bis[(dimethylamino-kappaN)methyl]-4-{3-[(S)-ethoxy(4-nitrophenoxy)phosphoryl]propyl}phenyl-kappaC~1~)(chloro)platinum(2+), CHLORIDE ION, Cutinase 1 | | Authors: | Rutten, L, Mannie, J.P.B.A, Lutz, M, Gros, P. | | Deposit date: | 2008-10-05 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Solid-state structural characterization of cutinase-ECE-pincer-metal hybrids
Chemistry, 15, 2009
|
|
6Z1Z
 
 | | Structure of the anti-CD9 nanobody 4C8 | | Descriptor: | Nanobody 4C8 | | Authors: | Neviani, N, Oosterheert, W, Pearce, N.M, Lutz, M, Kroon-Batenburg, L.M.J, Gros, P. | | Deposit date: | 2020-05-14 | | Release date: | 2020-09-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Implications for tetraspanin-enriched microdomain assembly based on structures of CD9 with EWI-F.
Life Sci Alliance, 3, 2020
|
|
2P8V
 
 | | Crystal structure of human Homer3 EVH1 domain | | Descriptor: | Homer protein homolog 3, SULFATE ION | | Authors: | Bouyain, S, Tu, J, Huang, G.N, Worley, P.F, Leahy, D. | | Deposit date: | 2007-03-23 | | Release date: | 2007-11-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | NFAT binding and regulation of T cell activation by the cytoplasmic scaffolding Homer proteins.
Science, 319, 2008
|
|
7ZNQ
 
 | | ABC transporter complex NosDFYL in GDN | | Descriptor: | COPPER (II) ION, Copper-binding lipoprotein NosL, MAGNESIUM ION, ... | | Authors: | Zhang, L, Mueller, C, Zipfel, S, Chami, M, Einsle, O. | | Deposit date: | 2022-04-21 | | Release date: | 2022-08-03 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Molecular interplay of an assembly machinery for nitrous oxide reductase.
Nature, 608, 2022
|
|
6Z1V
 
 | | Structure of the EC2 domain of CD9 in complex with nanobody 4E8 | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CD9 antigen, ... | | Authors: | Oosterheert, W, Pearce, N.M, Gros, P. | | Deposit date: | 2020-05-14 | | Release date: | 2020-09-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Implications for tetraspanin-enriched microdomain assembly based on structures of CD9 with EWI-F.
Life Sci Alliance, 3, 2020
|
|
6RLO
 
 | | Crystal structure of AT1412dm Fab fragment in complex with CD9 large extracellular loop | | Descriptor: | AT1412dm Fab Fragment (Heavy Chain), AT1412dm Fab Fragment (Light Chain), CD9 antigen, ... | | Authors: | Neviani, V, Pearce, N.M, Pos, W, Schotte, R, Spits, H, Gros, P. | | Deposit date: | 2019-05-02 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of a homo-dimerization site in tetraspanin CD9 targeted by a melanoma patient-derived antibody
To Be Published
|
|
6RLM
 
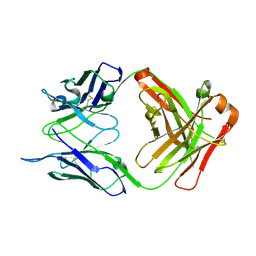 | | Crystal structure of AT1412dm Fab fragment | | Descriptor: | AT1412dm Fab (Heavy Chain), AT1412dm Fab (Light Chain), CHLORIDE ION | | Authors: | Neviani, V, Pearce, N.M, Pos, W, Schotte, R, Spits, H, Gros, P. | | Deposit date: | 2019-05-02 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of a homo-dimerization site in tetraspanin CD9 targeted by a melanoma patient-derived antibody
To Be Published
|
|
7AG5
 
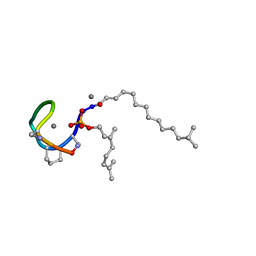 | | Structure of the Laspartomycin C double mutant G4D D-allo-Thr9D-Dap in complex with Geranyl phosphate | | Descriptor: | (~{E})-13-methyltetradec-2-enoic acid, CALCIUM ION, Geranyl phosphate, ... | | Authors: | Zeronian, M.R, Pearce, N.M, Wood, T.M, Martin, N.I, Janssen, B.J.C. | | Deposit date: | 2020-09-21 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Mechanistic insights into the C55-P targeting lipopeptide antibiotics revealed by structure-activity studies and high-resolution crystal structures
Chem Sci, 13, 2022
|
|
7O14
 
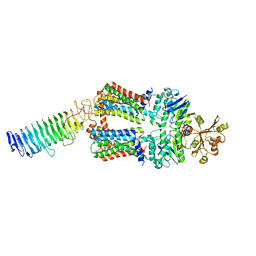 | | ABC transporter NosDFY, nucleotide-free in lipid nanodisc, R-domain 1 | | Descriptor: | MAGNESIUM ION, Probable ABC transporter ATP-binding protein NosF, Probable ABC transporter binding protein NosD, ... | | Authors: | Mueller, C, Zhang, L, Lu, W, Einsle, O, Du, J. | | Deposit date: | 2021-03-28 | | Release date: | 2022-04-13 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY | | Cite: | Molecular interplay of an assembly machinery for nitrous oxide reductase.
Nature, 608, 2022
|
|
7O15
 
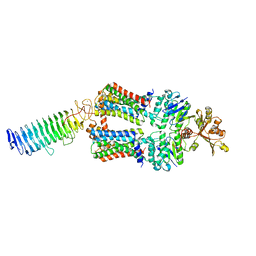 | | ABC transporter NosDFY, nucleotide-free in lipid nanodisc, R-domain 2 | | Descriptor: | MAGNESIUM ION, Probable ABC transporter ATP-binding protein NosF, Probable ABC transporter binding protein NosD, ... | | Authors: | Mueller, C, Zhang, L, Lu, W, Einsle, O, Du, J. | | Deposit date: | 2021-03-28 | | Release date: | 2022-04-13 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY | | Cite: | Molecular interplay of an assembly machinery for nitrous oxide reductase.
Nature, 608, 2022
|
|
7O12
 
 | | ABC transporter NosDFY, AMPPNP-bound in GDN | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Probable ABC transporter ATP-binding protein NosF, ... | | Authors: | Mueller, C, Zhang, L, Lu, W, Einsle, O, Du, J. | | Deposit date: | 2021-03-28 | | Release date: | 2022-04-13 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular interplay of an assembly machinery for nitrous oxide reductase.
Nature, 608, 2022
|
|
7O0Y
 
 | | ABC transporter NosDFY, nucleotide-free in GDN | | Descriptor: | MAGNESIUM ION, Probable ABC transporter ATP-binding protein NosF, Probable ABC transporter binding protein NosD, ... | | Authors: | Mueller, C, Zhang, L, Lu, W, Einsle, O, Du, J. | | Deposit date: | 2021-03-28 | | Release date: | 2022-04-13 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular interplay of an assembly machinery for nitrous oxide reductase.
Nature, 608, 2022
|
|
7O11
 
 | | ABC transporter NosDFY, nucleotide-free in GDN, R-domain 1 | | Descriptor: | MAGNESIUM ION, Probable ABC transporter ATP-binding protein NosF, Probable ABC transporter binding protein NosD, ... | | Authors: | Mueller, C, Zhang, L, Lu, W, Einsle, O, Du, J. | | Deposit date: | 2021-03-28 | | Release date: | 2022-04-13 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular interplay of an assembly machinery for nitrous oxide reductase.
Nature, 608, 2022
|
|
7O16
 
 | | ABC transporter NosDFY, nucleotide-free in lipid nanodisc, R-domain 3 | | Descriptor: | MAGNESIUM ION, Probable ABC transporter ATP-binding protein NosF, Probable ABC transporter binding protein NosD, ... | | Authors: | Mueller, C, Zhang, L, Lu, W, Einsle, O, Du, J. | | Deposit date: | 2021-03-28 | | Release date: | 2022-04-13 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY | | Cite: | Molecular interplay of an assembly machinery for nitrous oxide reductase.
Nature, 608, 2022
|
|
7O10
 
 | | ABC transporter NosDFY, nucleotide-free in GDN, R-domain 2 | | Descriptor: | MAGNESIUM ION, Probable ABC transporter ATP-binding protein NosF, Probable ABC transporter binding protein NosD, ... | | Authors: | Mueller, C, Zhang, L, Lu, W, Einsle, O, Du, J. | | Deposit date: | 2021-03-28 | | Release date: | 2022-04-13 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular interplay of an assembly machinery for nitrous oxide reductase.
Nature, 608, 2022
|
|
7O17
 
 | | ABC transporter NosDFY E154Q, ATP-bound in lipid nanodisc | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Probable ABC transporter ATP-binding protein NosF, ... | | Authors: | Mueller, C, Zhang, L, Lu, W, Einsle, O, Du, J. | | Deposit date: | 2021-03-28 | | Release date: | 2022-04-13 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Molecular interplay of an assembly machinery for nitrous oxide reductase.
Nature, 608, 2022
|
|
7O0Z
 
 | | ABC transporter NosFY, nucleotide-free in GDN | | Descriptor: | Probable ABC transporter ATP-binding protein NosF, Probable ABC transporter permease protein NosY | | Authors: | Mueller, C, Zhang, L, Lu, W, Einsle, O, Du, J. | | Deposit date: | 2021-03-28 | | Release date: | 2022-11-02 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular interplay of an assembly machinery for nitrous oxide reductase.
Nature, 608, 2022
|
|
