8AE4
 
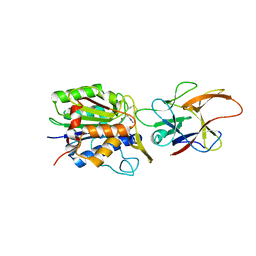 | | Crystal structure of human legumain in complex with Clitocypin 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Clitocypin-2, ... | | Authors: | Elamin, T, Brandstetter, H, Dall, E. | | Deposit date: | 2022-07-12 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural and functional studies of legumain-mycocypin complexes revealed a competitive, exosite-regulated mode of interaction.
J.Biol.Chem., 298, 2022
|
|
8AE5
 
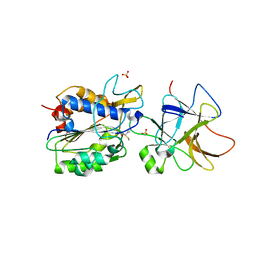 | | Crystal structure of human legumain in complex with macrocypin 1a | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Legumain, ... | | Authors: | Elamin, T, Brandstetter, H, Dall, E. | | Deposit date: | 2022-07-12 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural and functional studies of legumain-mycocypin complexes revealed a competitive, exosite-regulated mode of interaction.
J.Biol.Chem., 298, 2022
|
|
5I5L
 
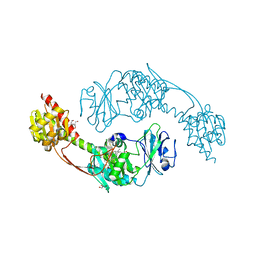 | | The photosensory module (PAS-GAF-PHY) of the bacterial phytochrome Agp1 (AtBphP1) in the Pr form, chromophore modelled with an endocyclic double bond in pyrrole ring A | | Descriptor: | BILIVERDINE IX ALPHA, Bacteriophytochrome protein, CALCIUM ION, ... | | Authors: | Nagano, S, Scheerer, P, Zubow, K, Lamparter, T, Krauss, N. | | Deposit date: | 2016-02-15 | | Release date: | 2016-08-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Crystal Structures of the N-terminal Photosensory Core Module of Agrobacterium Phytochrome Agp1 as Parallel and Anti-parallel Dimers.
J.Biol.Chem., 291, 2016
|
|
6R26
 
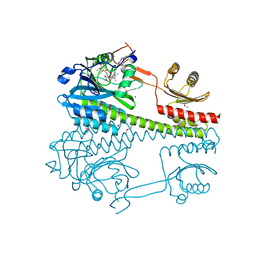 | | The photosensory core module (PAS-GAF-PHY) of the bacterial phytochrome Agp1 (AtBphP1) locked in a Pr-like state | | Descriptor: | 3-[2-[(~{Z})-[12-ethyl-6-(3-hydroxy-3-oxopropyl)-13-methyl-11-oxidanylidene-4,10-diazatricyclo[8.3.0.0^{3,7}]trideca-1,3,6,12-tetraen-5-ylidene]methyl]-5-[(~{Z})-(3-ethyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Bacteriophytochrome protein, CALCIUM ION | | Authors: | Scheerer, P, Michael, N, Lamparter, T, Krauss, N. | | Deposit date: | 2019-03-15 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Crystal structures of the photosensory core module of bacteriophytochrome Agp1 reveal pronounced structural flexibility of this protein in the red-absorbing Pr state
To Be Published
|
|
6R27
 
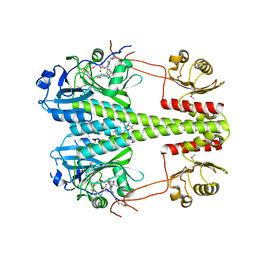 | | Crystallographic superstructure of the photosensory core module (PAS-GAF-PHY) of the bacterial phytochrome Agp1 (AtBphP1) locked in a Pr-like state | | Descriptor: | 3-[2-[(~{Z})-[12-ethyl-6-(3-hydroxy-3-oxopropyl)-13-methyl-11-oxidanylidene-4,10-diazatricyclo[8.3.0.0^{3,7}]trideca-1,3,6,12-tetraen-5-ylidene]methyl]-5-[(~{Z})-(3-ethyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Bacteriophytochrome protein | | Authors: | Scheerer, P, Michael, N, Lamparter, T, Krauss, N. | | Deposit date: | 2019-03-15 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structures of the photosensory core module of bacteriophytochrome Agp1 reveal pronounced structural flexibility of this protein in the red-absorbing Pr state
To Be Published
|
|
6R3U
 
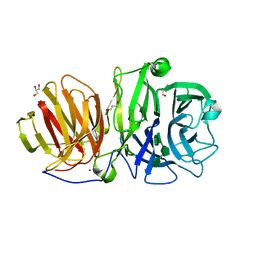 | | Endo-levanase BT1760 mutant E221A from Bacteroides thetaiotaomicron complexed with levantetraose | | Descriptor: | GLYCEROL, Glycoside hydrolase family 32, ZINC ION, ... | | Authors: | Eek, P, Ernits, K, Lukk, T, Alamae, T. | | Deposit date: | 2019-03-21 | | Release date: | 2019-06-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | First crystal structure of an endo-levanase - the BT1760 from a human gut commensal Bacteroides thetaiotaomicron.
Sci Rep, 9, 2019
|
|
6R3R
 
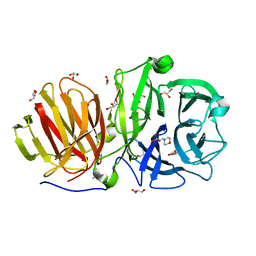 | | First crystal structure of endo-levanase BT1760 from Bacteroides thetaiotaomicron | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | Eek, P, Ernits, K, Lukk, T, Alamae, T. | | Deposit date: | 2019-03-21 | | Release date: | 2019-06-19 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | First crystal structure of an endo-levanase - the BT1760 from a human gut commensal Bacteroides thetaiotaomicron.
Sci Rep, 9, 2019
|
|
4UF8
 
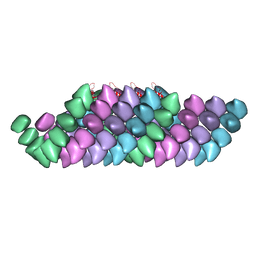 | | Electron cryo-microscopy structure of PB1-p62 filaments | | Descriptor: | SEQUESTOSOME-1 | | Authors: | Ciuffa, R, Lamark, T, Tarafder, A, Guesdon, A, Rybina, S, Hagen, W.J.H, Johansen, T, Sachse, C. | | Deposit date: | 2015-03-15 | | Release date: | 2015-05-13 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (10.9 Å) | | Cite: | The Selective Autophagy Receptor P62 Forms a Flexible Filamentous Helical Scaffold.
Cell Rep., 11, 2015
|
|
4UF9
 
 | | Electron cryo-microscopy structure of PB1-p62 type T filaments | | Descriptor: | SEQUESTOSOME-1 | | Authors: | Ciuffa, R, Lamark, T, Tarafder, A, Guesdon, A, Rybina, S, Hagen, W.J.H, Johansen, T, Sachse, C. | | Deposit date: | 2015-03-15 | | Release date: | 2015-05-13 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (10.3 Å) | | Cite: | The Selective Autophagy Receptor P62 Forms a Flexible Filamentous Helical Scaffold.
Cell Rep., 11, 2015
|
|
6YOO
 
 | | Structure of SAMM50 LIR bound to GABARAPL1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Gamma-aminobutyric acid receptor-associated protein-like 1, ... | | Authors: | Mouilleron, S, Wenxin, Z, Johansen, T, Tooze, S, Abudu, Y.P, Lamark, T. | | Deposit date: | 2020-04-14 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Structure of SAMM50 LIR bound to GABARAPL1
To Be Published
|
|
6YOP
 
 | | Structure of SAMM50 LIR bound to GABARAP | | Descriptor: | Sorting and assembly machinery component 50 homolog,Gamma-aminobutyric acid receptor-associated protein | | Authors: | Mouilleron, S, Wenxin, Z, Johansen, T, Tooze, S, Abudu, Y.P, Lamark, T. | | Deposit date: | 2020-04-14 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structure of SAMM50 LIR bound to GABARAP
To Be Published
|
|
5LXI
 
 | | GABARAP-L1 ATG4B LIR Complex | | Descriptor: | 1,2-ETHANEDIOL, Cysteine protease ATG4B, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Mouilleron, S, Skytte Rasmussen, M, Kumar Shrestha, B, Wirth, M, Bowitz Larsen, K, Abudu Princely, Y, Sjottem, E, Tooze, S, Lamark, T, Johansen, T, Lee, R. | | Deposit date: | 2016-09-21 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | ATG4B contains a C-terminal LIR motif important for binding and efficient cleavage of mammalian orthologs of yeast Atg8.
Autophagy, 13, 2017
|
|
5LXH
 
 | | GABARAP-L1 ATG4B LIR Complex | | Descriptor: | Cysteine protease ATG4B, GLYCEROL, Gamma-aminobutyric acid receptor-associated protein-like 1, ... | | Authors: | Mouilleron, S, Skytte Rasmussen, M, Kumar Shrestha, B, Wirth, M, Bowitz Larsen, K, Abudu Princely, Y, Sjottem, E, Tooze, S, Lamark, T, Johansen, T, Lee, R. | | Deposit date: | 2016-09-21 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | ATG4B contains a C-terminal LIR motif important for binding and efficient cleavage of mammalian orthologs of yeast Atg8.
Autophagy, 13, 2017
|
|
4U63
 
 | | Crystal structure of a bacterial class III photolyase from Agrobacterium tumefaciens at 1.67A resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5,10-METHENYL-6,7,8-TRIHYDROFOLIC ACID, DNA photolyase, ... | | Authors: | Scheerer, P, Zhang, F, Kalms, J, von Stetten, D, Krauss, N, Oberpichler, I, Lamparter, T. | | Deposit date: | 2014-07-26 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | The Class III Cyclobutane Pyrimidine Dimer Photolyase Structure Reveals a New Antenna Chromophore Binding Site and Alternative Photoreduction Pathways.
J.Biol.Chem., 290, 2015
|
|
5KCM
 
 | | Crystal structure of iron-sulfur cluster containing photolyase PhrB mutant I51W | | Descriptor: | (6-4) photolyase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Yang, X, Bowatte, K, Zhang, F, Lamparter, T. | | Deposit date: | 2016-06-06 | | Release date: | 2017-01-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.149 Å) | | Cite: | Crystal Structures of Bacterial (6-4) Photolyase Mutants with Impaired DNA Repair Activity.
Photochem. Photobiol., 93, 2017
|
|
5LFA
 
 | | Crystal structure of iron-sulfur cluster containing bacterial (6-4) photolyase PhrB - Y424F mutant with impaired DNA repair activity | | Descriptor: | (6-4) photolyase, 1-deoxy-1-(6,7-dimethyl-2,4-dioxo-3,4-dihydropteridin-8(2H)-yl)-D-ribitol, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kwiatkowski, D, Zhang, F, Krauss, N, Lamparter, T, Scheerer, P. | | Deposit date: | 2016-06-30 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of Bacterial (6-4) Photolyase Mutants with Impaired DNA Repair Activity.
Photochem. Photobiol., 93, 2017
|
|
2W26
 
 | | Factor Xa in complex with BAY59-7939 | | Descriptor: | 5-chloro-N-({(5S)-2-oxo-3-[4-(3-oxomorpholin-4-yl)phenyl]-1,3-oxazolidin-5-yl}methyl)thiophene-2-carboxamide, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION | | Authors: | Roehrig, S, Straub, A, Pohlmann, J, Lampe, T, Pernerstorfer, J, Schlemmer, K, Reinemer, P, Perzborn, E, Schaefer, M. | | Deposit date: | 2008-10-24 | | Release date: | 2008-11-11 | | Last modified: | 2021-04-28 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Discovery of the Novel Antithrombotic Agent 5-Chloro-N-({(5S)-2-Oxo-3- [4-(3-Oxomorpholin-4-Yl)Phenyl]-1,3-Oxazolidin-5-Yl}Methyl)Thiophene-2- Carboxamide (Bay 59-7939): An Oral, Direct Factor Xa Inhibitor.
J.Med.Chem., 48, 2005
|
|
6HOG
 
 | | Structure of VPS34 LIR motif bound to GABARAP | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Phosphatidylinositol 3-kinase catalytic subunit type 3,Gamma-aminobutyric acid receptor-associated protein, ... | | Authors: | Mouilleron, S, Birgisdottir, A.B, Bhujbal, Z, Wirth, M, Sjottem, E, Evjen, G, Zhang, W, Lee, R, O'Reilly, N, Tooze, S, Lamark, T, Johansen, T. | | Deposit date: | 2018-09-17 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Members of the autophagy class III phosphatidylinositol 3-kinase complex I interact with GABARAP and GABARAPL1 via LIR motifs.
Autophagy, 15, 2019
|
|
6HOK
 
 | | Structure of Beclin1 LIR (S96E) motif bound to GABARAP | | Descriptor: | 1,2-ETHANEDIOL, Beclin-1,Gamma-aminobutyric acid receptor-associated protein | | Authors: | Mouilleron, S, Birgisdottir, A.B, Bhujbal, Z, Wirth, M, Sjottem, E, Evjen, G, Zhang, W, Lee, R, O'Reilly, N, Tooze, S, Lamark, T, Johansen, T. | | Deposit date: | 2018-09-17 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Members of the autophagy class III phosphatidylinositol 3-kinase complex I interact with GABARAP and GABARAPL1 via LIR motifs.
Autophagy, 15, 2019
|
|
6HOL
 
 | | Structure of ATG14 LIR motif bound to GABARAPL1 | | Descriptor: | Beclin 1-associated autophagy-related key regulator, GLYCEROL, Gamma-aminobutyric acid receptor-associated protein-like 1, ... | | Authors: | Mouilleron, S, Birgisdottir, A.B, Bhujbal, Z, Wirth, M, Sjottem, E, Evjen, G, Zhang, W, Lee, R, O'Reilly, N, Tooze, S, Lamark, T, Johansen, T. | | Deposit date: | 2018-09-17 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Members of the autophagy class III phosphatidylinositol 3-kinase complex I interact with GABARAP and GABARAPL1 via LIR motifs.
Autophagy, 15, 2019
|
|
6HOJ
 
 | | Structure of Beclin1 LIR motif bound to GABARAP | | Descriptor: | 1,2-ETHANEDIOL, Beclin-1,Gamma-aminobutyric acid receptor-associated protein, SULFATE ION | | Authors: | Mouilleron, S, Birgisdottir, A.B, Bhujbal, Z, Wirth, M, Sjottem, E, Evjen, G, Zhang, W, Lee, R, O'Reilly, N, Tooze, S, Lamark, T, Johansen, T. | | Deposit date: | 2018-09-17 | | Release date: | 2019-02-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Members of the autophagy class III phosphatidylinositol 3-kinase complex I interact with GABARAP and GABARAPL1 via LIR motifs.
Autophagy, 15, 2019
|
|
6HOI
 
 | | Structure of Beclin1 LIR motif bound to GABARAPL1 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Beclin-1, ... | | Authors: | Mouilleron, S, Birgisdottir, A.B, Bhujbal, Z, Wirth, M, Sjottem, E, Evjen, G, Zhang, W, Lee, R, O'Reilly, N, Tooze, S, Lamark, T, Johansen, T. | | Deposit date: | 2018-09-17 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Members of the autophagy class III phosphatidylinositol 3-kinase complex I interact with GABARAP and GABARAPL1 via LIR motifs.
Autophagy, 15, 2019
|
|
6HOH
 
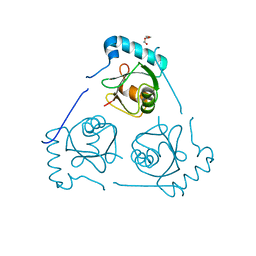 | | Structure of VPS34 LIR motif (S249E) bound to GABARAP | | Descriptor: | Phosphatidylinositol 3-kinase catalytic subunit type 3,Gamma-aminobutyric acid receptor-associated protein, TRIETHYLENE GLYCOL | | Authors: | Mouilleron, S, Birgisdottir, A.B, Bhujbal, Z, Wirth, M, Sjottem, E, Evjen, G, Zhang, W, Lee, R, O'Reilly, N, Tooze, S, Lamark, T, Johansen, T. | | Deposit date: | 2018-09-17 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Members of the autophagy class III phosphatidylinositol 3-kinase complex I interact with GABARAP and GABARAPL1 via LIR motifs.
Autophagy, 15, 2019
|
|
2G5L
 
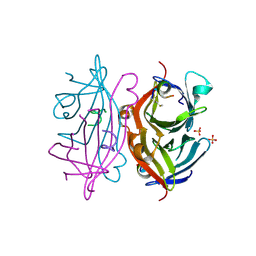 | | Streptavidin in complex with Nanotag | | Descriptor: | (FME)(ASP)(VAL)(GLU)(ALA)(TRP)(LEU), GLYCEROL, SULFATE ION, ... | | Authors: | Perbandt, M, Bruns, O, Vallazza, M, Lamla, T, Betzel, C, Erdmann, V.A. | | Deposit date: | 2006-02-23 | | Release date: | 2007-02-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | High resolution structure of Streptavidin in complex with a novel high affinity peptide tag mimicking the biotin binding motif
To be Published
|
|
4DJA
 
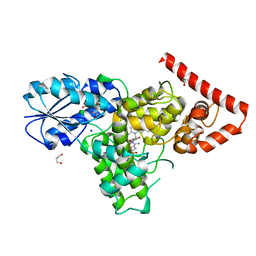 | | Crystal structure of a prokaryotic (6-4) photolyase PhrB from Agrobacterium Tumefaciens with an Fe-S cluster and a 6,7-dimethyl-8-ribityllumazine antenna chromophore at 1.45A resolution | | Descriptor: | 1-deoxy-1-(6,7-dimethyl-2,4-dioxo-3,4-dihydropteridin-8(2H)-yl)-D-ribitol, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Scheerer, P, Zhang, F, Oberpichler, I, Lamparter, T, Krauss, N. | | Deposit date: | 2012-02-01 | | Release date: | 2013-04-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of a prokaryotic (6-4) photolyase with an Fe-S cluster and a 6,7-dimethyl-8-ribityllumazine antenna chromophore.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
