2JR3
 
 | | Antibacterial Peptide from Eggshell Matrix: Structure and Self-assembly of beta-defensin Like Peptide from the Chinese Soft-shelled Turtle Eggshell | | Descriptor: | Pelovaterin | | Authors: | Vivekanandan, S, Lakshminarayanan, R, Jois, S.D.S, Perumal Samy, R, Banerjee, Y, Chi-Jin, E.O, Teo, K.W, Kini, R.M, Valiyaveettil, S. | | Deposit date: | 2007-06-20 | | Release date: | 2008-05-27 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Structure, self-assembly, and dual role of a beta-defensin-like peptide from the Chinese soft-shelled turtle eggshell matrix.
J.Am.Chem.Soc., 130, 2008
|
|
4BEA
 
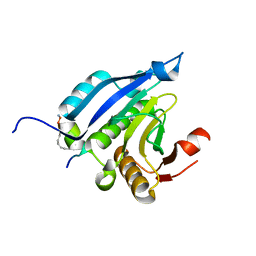 | | Crystal Structure of eIF4E in Complex with a Stapled Peptide Derivative | | Descriptor: | Eukaryotic translation initiation factor 4E, STAPLED EIF4E INTERACTING PEPTIDE | | Authors: | Quah, S.T, Lama, D, Verma, C.S, Lane, D.P, Brown, C.J. | | Deposit date: | 2013-03-07 | | Release date: | 2013-12-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Rational Optimization of Conformational Effects Induced by Hydrocarbon Staples in Peptides and Their Binding Interfaces.
Sci.Rep., 3, 2013
|
|
4UD7
 
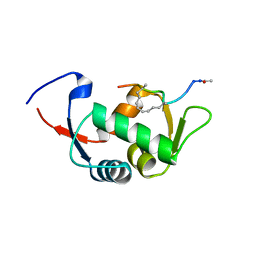 | | Structure of the stapled peptide YS-02 bound to MDM2 | | Descriptor: | MDM2, YS-02 | | Authors: | Tan, Y.S, Reeks, J, Brown, C.J, Jennings, C.E, Eapen, R.S, Tng, Q.S, Thean, D, Ying, Y.T, Gago, F.J.F, Lane, D.P, Noble, M.E.M, Verma, C. | | Deposit date: | 2014-12-08 | | Release date: | 2016-01-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Benzene Probes in Molecular Dynamics Simulations Reveal Novel Binding Sites for Ligand Design.
J Phys Chem Lett, 7, 2016
|
|
4UE1
 
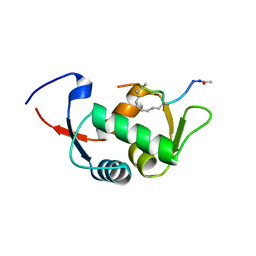 | | Structure of the stapled peptide YS-01 bound to MDM2 | | Descriptor: | E3 UBIQUITIN-PROTEIN LIGASE MDM2, YS-01 | | Authors: | Tan, Y.S, Reeks, J, Brown, C.J, Jennings, C.E, Eapen, R.S, Tng, Q.S, Thean, D, Ying, Y.T, Gago, F.J.F, Lane, D.P, Noble, M.E.M, Verma, C. | | Deposit date: | 2014-12-14 | | Release date: | 2016-01-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Benzene Probes in Molecular Dynamics Simulations Reveal Novel Binding Sites for Ligand Design.
J Phys Chem Lett, 7, 2016
|
|
