4OK7
 
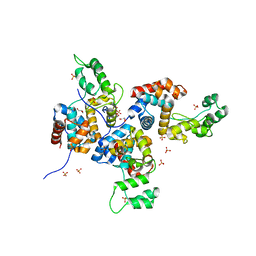 | | Structure of bacteriophage SPN1S endolysin from Salmonella typhimurium | | Descriptor: | Endolysin, GLYCEROL, SULFATE ION | | Authors: | Park, Y, Lim, J, Kong, M, Ryu, S, Rhee, S. | | Deposit date: | 2014-01-22 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of bacteriophage SPN1S endolysin reveals an unusual two-module fold for the peptidoglycan lytic and binding activity.
Mol.Microbiol., 92, 2014
|
|
6ILU
 
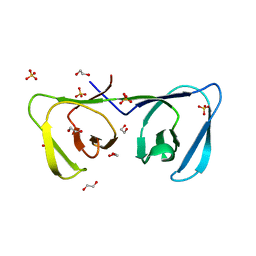 | | Endolysin LysPBC5 CBD | | Descriptor: | 1,2-ETHANEDIOL, Lysin, SULFATE ION | | Authors: | Suh, J.Y, Ryu, K.S, Ryu, S, Lee, K.O, Kong, M.S, Bae, J.W, Kim, I.T. | | Deposit date: | 2018-10-19 | | Release date: | 2019-07-31 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Structural Basis for Cell-Wall Recognition by Bacteriophage PBC5 Endolysin.
Structure, 27, 2019
|
|
4IYP
 
 | | structure of the nPP2Ac-alpha4 complex | | Descriptor: | Immunoglobulin-binding protein 1, Serine/threonine-protein phosphatase 2A catalytic subunit alpha isoform | | Authors: | Jiang, L, Stanevich, V, Satyshur, K.A, Xing, Y. | | Deposit date: | 2013-01-29 | | Release date: | 2013-04-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.797 Å) | | Cite: | Structural basis of protein phosphatase 2A stable latency.
Nat Commun, 4, 2013
|
|
