1SKY
 
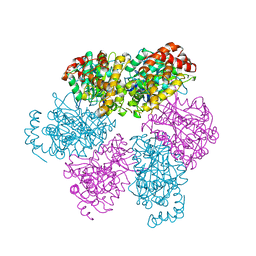 | | CRYSTAL STRUCTURE OF THE NUCLEOTIDE FREE ALPHA3BETA3 SUB-COMPLEX OF F1-ATPASE FROM THE THERMOPHILIC BACILLUS PS3 | | Descriptor: | F1-ATPASE, SULFATE ION | | Authors: | Shirakihara, Y, Leslie, A.G.W, Abrahams, J.P, Walker, J.E, Ueda, T, Sekimoto, Y, Kambara, M, Saika, K, Kagawa, Y, Yoshida, M. | | Deposit date: | 1997-02-26 | | Release date: | 1998-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The crystal structure of the nucleotide-free alpha 3 beta 3 subcomplex of F1-ATPase from the thermophilic Bacillus PS3 is a symmetric trimer.
Structure, 5, 1997
|
|
4XD7
 
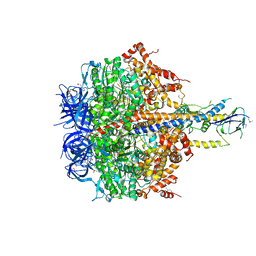 | | Structure of thermophilic F1-ATPase inhibited by epsilon subunit | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP synthase epsilon chain, ATP synthase gamma chain, ... | | Authors: | SHIRAKIHARA, Y, SHIRATORI, A, TANIKAWA, H, NAKASAKO, M, YOSHIDA, M, SUZUKI, T. | | Deposit date: | 2014-12-19 | | Release date: | 2015-08-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structure of a thermophilic F1 -ATPase inhibited by an epsilon-subunit: deeper insight into the epsilon-inhibition mechanism.
Febs J., 282, 2015
|
|
1PFK
 
 | |
7TD0
 
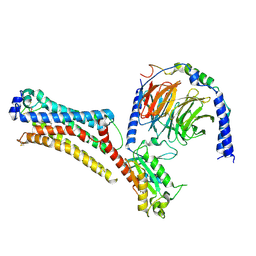 | | Lysophosphatidic acid receptor 1-Gi complex bound to LPA | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, S, Paknejad, N, Zhu, L, Kihara, Y, Ray, D, Chun, J, Liu, W, Hite, R.K, Huang, X.Y. | | Deposit date: | 2021-12-30 | | Release date: | 2022-02-09 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Differential activation mechanisms of lipid GPCRs by lysophosphatidic acid and sphingosine 1-phosphate.
Nat Commun, 13, 2022
|
|
7TD2
 
 | | Lysophosphatidic acid receptor 1-Gi complex bound to LPA, state a | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, S, Paknejad, N, Zhu, L, Kihara, Y, Ray, D, Chun, J, Liu, W, Hite, R.K, Huang, X.Y. | | Deposit date: | 2021-12-30 | | Release date: | 2022-02-09 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Differential activation mechanisms of lipid GPCRs by lysophosphatidic acid and sphingosine 1-phosphate.
Nat Commun, 13, 2022
|
|
7TD3
 
 | | Sphingosine-1-phosphate receptor 1-Gi complex bound to S1P | | Descriptor: | (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liu, S, Paknejad, N, Zhu, L, Kihara, Y, Ray, D, Chun, J, Liu, W, Hite, R.K, Huang, X.Y. | | Deposit date: | 2021-12-30 | | Release date: | 2022-02-09 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Differential activation mechanisms of lipid GPCRs by lysophosphatidic acid and sphingosine 1-phosphate.
Nat Commun, 13, 2022
|
|
7TD1
 
 | | Lysophosphatidic acid receptor 1-Gi complex bound to LPA, state a | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, S, Paknejad, N, Zhu, L, Kihara, Y, Ray, D, Chun, J, Liu, W, Hite, R.K, Huang, X.Y. | | Deposit date: | 2021-12-30 | | Release date: | 2022-02-09 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Differential activation mechanisms of lipid GPCRs by lysophosphatidic acid and sphingosine 1-phosphate.
Nat Commun, 13, 2022
|
|
7TD4
 
 | | Sphingosine-1-phosphate receptor 1-Gi complex bound to Siponimod | | Descriptor: | 1-[[4-[(~{E})-~{N}-[[4-cyclohexyl-3-(trifluoromethyl)phenyl]methoxy]-~{C}-methyl-carbonimidoyl]-2-ethyl-phenyl]methyl]azetidine-3-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liu, S, Paknejad, N, Zhu, L, Kihara, Y, Ray, D, Chun, J, Liu, W, Hite, R.K, Huang, X.Y. | | Deposit date: | 2021-12-30 | | Release date: | 2022-02-09 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Differential activation mechanisms of lipid GPCRs by lysophosphatidic acid and sphingosine 1-phosphate.
Nat Commun, 13, 2022
|
|
4Z34
 
 | | Crystal Structure of Human Lysophosphatidic Acid Receptor 1 in complex with ONO9780307 | | Descriptor: | (2S)-2,3-dihydroxypropyl (7Z)-tetradec-7-enoate, Lysophosphatidic acid receptor 1, Soluble cytochrome b562, ... | | Authors: | Chrencik, J.E, Roth, C.B, Terakado, M, Kurata, H, Omi, R, Kihara, Y, Warshaviak, D, Nakade, S, Asmar-Rovira, G, Mileni, M, Mizuno, H, Griffith, M.T, Rodgers, C, Han, G.W, Velasquez, J, Chun, J, Stevens, R.C, Hanson, M.A, GPCR Network (GPCR) | | Deposit date: | 2015-03-30 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of Antagonist Bound Human Lysophosphatidic Acid Receptor 1.
Cell, 161, 2015
|
|
4Z35
 
 | | Crystal Structure of Human Lysophosphatidic Acid Receptor 1 in complex with ONO-9910539 | | Descriptor: | (2S)-2,3-dihydroxypropyl (7Z)-tetradec-7-enoate, 3-{1-[(2S,3S)-3-(4-acetyl-3,5-dimethoxyphenyl)-2-(2,3-dihydro-1H-inden-2-ylmethyl)-3-hydroxypropyl]-4-(methoxycarbonyl)-1H-pyrrol-3-yl}propanoic acid, Lysophosphatidic acid receptor 1,Soluble cytochrome b562 | | Authors: | Chrencik, J.E, Roth, C.B, Terakado, M, Kurata, H, Omi, R, Kihara, Y, Warshaviak, D, Nakade, S, Asmar-Rovira, G, Mileni, M, Mizuno, H, Griffith, M.T, Rodgers, C, Han, G.W, Velasquez, J, Chun, J, Stevens, R.C, Hanson, M.A, GPCR Network (GPCR) | | Deposit date: | 2015-03-30 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of Antagonist Bound Human Lysophosphatidic Acid Receptor 1.
Cell, 161, 2015
|
|
4Z36
 
 | | Crystal Structure of Human Lysophosphatidic Acid Receptor 1 in complex with ONO-3080573 | | Descriptor: | (2S)-2,3-dihydroxypropyl (7Z)-tetradec-7-enoate, 1-(4-{[(2S,3R)-2-(2,3-dihydro-1H-inden-2-yloxy)-3-(3,5-dimethoxy-4-methylphenyl)-3-hydroxypropyl]oxy}phenyl)cyclopropanecarboxylic acid, Lysophosphatidic acid receptor 1,Soluble cytochrome b562 | | Authors: | Chrencik, J.E, Roth, C.B, Terakado, M, Kurata, H, Omi, R, Kihara, Y, Warshaviak, D, Nakade, S, Asmar-Rovira, G, Mileni, M, Mizuno, H, Griffith, M.T, Rodgers, C, Han, G.W, Velasquez, J, Chun, J, Stevens, R.C, Hanson, M.A, GPCR Network (GPCR) | | Deposit date: | 2015-03-30 | | Release date: | 2015-06-03 | | Last modified: | 2015-07-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of Antagonist Bound Human Lysophosphatidic Acid Receptor 1.
Cell, 161, 2015
|
|
3X37
 
 | | Crystal structure of the N-terminal domain of Sld7 in complex with Sld3 | | Descriptor: | GLYCEROL, Mitochondrial morphogenesis protein SLD7, ZYRO0C14696p | | Authors: | Itou, H, Araki, H, Shirakihara, Y. | | Deposit date: | 2015-01-16 | | Release date: | 2015-08-19 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The quaternary structure of the eukaryotic DNA replication proteins Sld7 and Sld3.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3X38
 
 | | Crystal structure of the C-terminal domain of Sld7 | | Descriptor: | GLYCEROL, Mitochondrial morphogenesis protein SLD7, SULFATE ION | | Authors: | Itou, H, Araki, H, Shirakihara, Y. | | Deposit date: | 2015-01-16 | | Release date: | 2015-08-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | The quaternary structure of the eukaryotic DNA replication proteins Sld7 and Sld3.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3IF5
 
 | | Crystal Structure Analysis of Mglu | | Descriptor: | Salt-tolerant glutaminase | | Authors: | Yoshimune, K, Shirakihara, Y. | | Deposit date: | 2009-07-24 | | Release date: | 2009-08-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Crystal structure of salt-tolerant glutaminase from Micrococcus luteus K-3 in the presence and absence of its product L-glutamate and its activator Tris.
Febs J., 277, 2010
|
|
3IHA
 
 | | Crystal Structure Analysis of Mglu in its glutamate form | | Descriptor: | GLUTAMIC ACID, Salt-tolerant glutaminase | | Authors: | Yoshimune, K, Shirakihara, Y. | | Deposit date: | 2009-07-29 | | Release date: | 2010-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of salt-tolerant glutaminase from Micrococcus luteus K-3 in the presence and absence of its product l-glutamate and its activator Tris
Febs J., 277, 2010
|
|
3IH8
 
 | | Crystal Structure Analysis of Mglu in its native form | | Descriptor: | Salt-tolerant glutaminase | | Authors: | Yoshimune, K, Shirakihara, Y. | | Deposit date: | 2009-07-29 | | Release date: | 2010-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of salt-tolerant glutaminase from Micrococcus luteus K-3 in the presence and absence of its product l-glutamate and its activator Tris
Febs J., 277, 2010
|
|
3IH9
 
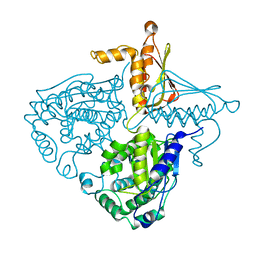 | | Crystal Structure Analysis of Mglu in its tris form | | Descriptor: | Salt-tolerant glutaminase | | Authors: | Yoshimune, K, Shirakihara, Y. | | Deposit date: | 2009-07-29 | | Release date: | 2010-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of salt-tolerant glutaminase from Micrococcus luteus K-3 in the presence and absence of its product l-glutamate and its activator Tris
Febs J., 277, 2010
|
|
3IHB
 
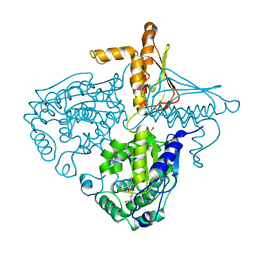 | | Crystal Structure Analysis of Mglu in its tris and glutamate form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLUTAMIC ACID, Salt-tolerant glutaminase | | Authors: | Yoshimune, K, Shirakihara, Y. | | Deposit date: | 2009-07-29 | | Release date: | 2010-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of salt-tolerant glutaminase from Micrococcus luteus K-3 in the presence and absence of its product l-glutamate and its activator Tris
Febs J., 277, 2010
|
|
1IYX
 
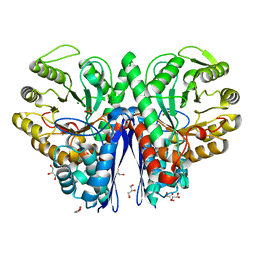 | | Crystal structure of enolase from Enterococcus hirae | | Descriptor: | ENOLASE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Hosaka, T, Meguro, T, Yamato, I, Shirakihara, Y. | | Deposit date: | 2002-09-12 | | Release date: | 2003-07-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Enterococcus hirae Enolase at 2.8 A Resolution
J.BIOCHEM.(TOKYO), 133, 2003
|
|
2YVE
 
 | | Crystal structure of the methylene blue-bound form of the multi-drug binding transcriptional repressor CgmR | | Descriptor: | 3,7-BIS(DIMETHYLAMINO)PHENOTHIAZIN-5-IUM, CHLORIDE ION, GLYCEROL, ... | | Authors: | Itou, H, Shirakihara, Y, Tanaka, I. | | Deposit date: | 2007-04-12 | | Release date: | 2008-04-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structures of the Multidrug Binding Repressor Corynebacteriumglutamicum CgmR in Complex with Inducers and with an Operator
J.Mol.Biol., 403, 2010
|
|
2YVH
 
 | |
2ZOZ
 
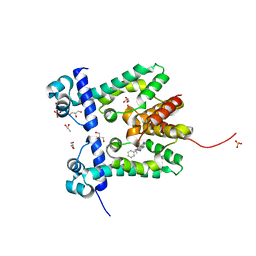 | | Crystal structure of the ethidium-bound form of the multi-drug binding transcriptional repressor CgmR | | Descriptor: | ETHIDIUM, GLYCEROL, SULFATE ION, ... | | Authors: | Itou, H, Shirakihara, Y, Tanaka, I. | | Deposit date: | 2008-06-20 | | Release date: | 2008-07-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structures of the Multidrug Binding Repressor Corynebacteriumglutamicum CgmR in Complex with Inducers and with an Operator
J.Mol.Biol., 403, 2010
|
|
2Z5H
 
 | | Crystal structure of the head-to-tail junction of tropomyosin complexed with a fragment of TnT | | Descriptor: | General control protein GCN4 and Tropomyosin alpha-1 chain, Tropomyosin alpha-1 chain and General control protein GCN4, Troponin T, ... | | Authors: | Murakami, K, Nozawa, K, Tomii, K, Kudou, N, Igarashi, N, Shirakihara, Y, Wakatsuki, S, Stewart, M, Yasunaga, T, Wakabayashi, T. | | Deposit date: | 2007-07-12 | | Release date: | 2008-04-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structural basis for tropomyosin overlap in thin (actin) filaments and the generation of a molecular swivel by troponin-T
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2Z5I
 
 | | Crystal structure of the head-to-tail junction of tropomyosin | | Descriptor: | General control protein GCN4 and Tropomyosin alpha-1 chain, MAGNESIUM ION, Tropomyosin alpha-1 chain and General control protein GCN4 | | Authors: | Murakami, K, Nozawa, K, Tomii, K, Kudou, N, Igarashi, N, Shirakihara, Y, Wakatsuki, S, Stewart, M, Yasunaga, T, Wakabayashi, T. | | Deposit date: | 2007-07-12 | | Release date: | 2008-04-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for tropomyosin overlap in thin (actin) filaments and the generation of a molecular swivel by troponin-T
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3AGE
 
 | |
