2AY7
 
 | |
2AY5
 
 | |
2AY2
 
 | |
2AY9
 
 | |
2AY3
 
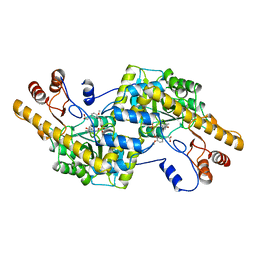 | | AROMATIC AMINO ACID AMINOTRANSFERASE WITH 3-(3,4-DIMETHOXYPHENYL)PROPIONIC ACID | | Descriptor: | 3-(3,4-DIMETHOXYPHENYL)PROPIONIC ACID, AROMATIC AMINO ACID AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Okamoto, A, Hirotsu, K, Kagamiyama, H. | | Deposit date: | 1998-08-06 | | Release date: | 1999-02-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The active site of Paracoccus denitrificans aromatic amino acid aminotransferase has contrary properties: flexibility and rigidity.
Biochemistry, 38, 1999
|
|
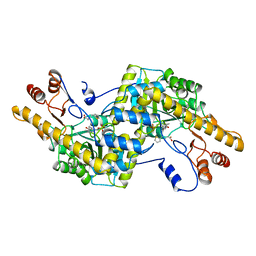
2AY8
 
 | |
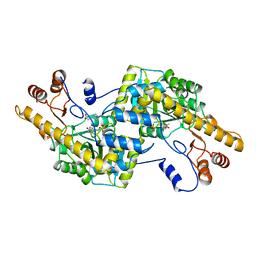
2AY1
 
 | |
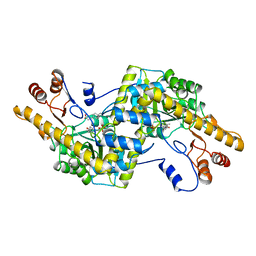
2AY4
 
 | |
2AY6
 
 | |
1ART
 
 | |
1AY4
 
 | | AROMATIC AMINO ACID AMINOTRANSFERASE WITHOUT SUBSTRATE | | Descriptor: | AROMATIC AMINO ACID AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Okamoto, A, Hirotsu, K, Kagamiyama, H. | | Deposit date: | 1997-11-14 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structures of Paracoccus denitrificans aromatic amino acid aminotransferase: a substrate recognition site constructed by rearrangement of hydrogen bond network.
J.Mol.Biol., 280, 1998
|
|
1AY5
 
 | | AROMATIC AMINO ACID AMINOTRANSFERASE COMPLEX WITH MALEATE | | Descriptor: | AROMATIC AMINO ACID AMINOTRANSFERASE, MALEIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Okamoto, A, Hirotsu, K, Kagamiyama, H. | | Deposit date: | 1997-11-14 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of Paracoccus denitrificans aromatic amino acid aminotransferase: a substrate recognition site constructed by rearrangement of hydrogen bond network.
J.Mol.Biol., 280, 1998
|
|
9IZ4
 
 | |
9IZ3
 
 | |
7CBD
 
 | | Catalytic domain of Cellulomonas fimi Cel6B | | Descriptor: | Exoglucanase A | | Authors: | Nakamura, A, Ishiwata, D, Visootsat, A, Uchiyama, T, Mizutani, K, Kaneko, S, Murata, T, Igarashi, K, Iino, R. | | Deposit date: | 2020-06-12 | | Release date: | 2020-08-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Domain architecture divergence leads to functional divergence in binding and catalytic domains of bacterial and fungal cellobiohydrolases.
J.Biol.Chem., 295, 2020
|
|
1AY8
 
 | |
1ARS
 
 | |
3VM6
 
 | | Crystal structure of ribose-1,5-bisphosphate isomerase from Thermococcus kodakarensis KOD1 in complex with alpha-D-ribose-1,5-bisphosphate | | Descriptor: | 1,5-di-O-phosphono-alpha-D-ribofuranose, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Nakamura, A, Fujihashi, M, Aono, R, Sato, T, Nishiba, Y, Yoshida, S, Yano, A, Atomi, H, Imanaka, T, Miki, K. | | Deposit date: | 2011-12-08 | | Release date: | 2012-04-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Dynamic, ligand-dependent conformational change triggers reaction of ribose-1,5-bisphosphate isomerase from Thermococcus kodakarensis KOD1
J.Biol.Chem., 287, 2012
|
|
3IP4
 
 | | The high resolution structure of GatCAB | | Descriptor: | Aspartyl/glutamyl-tRNA(Asn/Gln) amidotransferase subunit B, Aspartyl/glutamyl-tRNA(Asn/Gln) amidotransferase subunit C, Glutamyl-tRNA(Gln) amidotransferase subunit A, ... | | Authors: | Nakamura, A, Yao, M, Tanaka, I. | | Deposit date: | 2009-08-17 | | Release date: | 2009-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Two distinct regions in Staphylococcus aureus GatCAB guarantee accurate tRNA recognition
Nucleic Acids Res., 38, 2010
|
|
8GCL
 
 | | Cryo-EM structure of hAQP2 in DDM | | Descriptor: | Aquaporin-2 | | Authors: | Kamegawa, A, Suzuki, S, Nishikawa, K, Numoto, N, Suzuki, H, Fujiyoshi, Y. | | Deposit date: | 2023-03-02 | | Release date: | 2023-06-21 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structural analysis of the water channel AQP2 by single-particle cryo-EM.
J.Struct.Biol., 215, 2023
|
|
8HRF
 
 | |
4ZHR
 
 | | Structure of HIV-1 RT Q151M mutant | | Descriptor: | RT p51 subunit, RT p66 subunit | | Authors: | Nakamura, A, Tamura, N, Yasutake, Y. | | Deposit date: | 2015-04-27 | | Release date: | 2015-11-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structure of the HIV-1 reverse transcriptase Q151M mutant: insights into the inhibitor resistance of HIV-1 reverse transcriptase and the structure of the nucleotide-binding pocket of Hepatitis B virus polymerase.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
4ZM7
 
 | |
5Z7M
 
 | | SmChiA sliding-intermediate with chitohexaose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, Chitinase A, ... | | Authors: | Nakamura, A, Iino, R. | | Deposit date: | 2018-01-30 | | Release date: | 2018-09-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Processive chitinase is Brownian monorail operated by fast catalysis after peeling rail from crystalline chitin.
Nat Commun, 9, 2018
|
|
5Z7N
 
 | | SmChiA sliding-intermediate with chitopentaose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, Chitinase A, ... | | Authors: | Nakamura, A, Iino, R. | | Deposit date: | 2018-01-30 | | Release date: | 2018-09-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Processive chitinase is Brownian monorail operated by fast catalysis after peeling rail from crystalline chitin.
Nat Commun, 9, 2018
|
|
