1D7N
 
 | | SOLUTION STRUCTURE ANALYSIS OF THE MASTOPARAN WITH DETERGENTS | | Descriptor: | PROTEIN (WASP VENOM PEPTIDE (MASTOPARAN)) | | Authors: | Hori, Y, Demura, M, Iwadate, M, Niidome, T, Aoyagi, H, Asakura, T. | | Deposit date: | 1999-10-19 | | Release date: | 2001-06-20 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Interaction of mastoparan with membranes studied by 1H-NMR spectroscopy in detergent micelles and by solid-state 2H-NMR and 15N-NMR spectroscopy in oriented lipid bilayers.
Eur.J.Biochem., 268, 2001
|
|
1WMX
 
 | | Crystal Structure of Family 30 Carbohydrate Binding Module | | Descriptor: | COG3291: FOG: PKD repeat, SULFATE ION | | Authors: | Horiguchi, Y, Kono, M, Suzuki, A, Yamane, T, Arai, M, Sakka, K, Omiya, K. | | Deposit date: | 2004-07-21 | | Release date: | 2004-08-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Family 30 Carbohydrate Binding Module
To be Published
|
|
1WZX
 
 | | Crystal Structure of Family 30 Carbohydrate Binding Module. | | Descriptor: | COG3291: FOG: PKD repeat | | Authors: | Horiguchi, Y, Kono, M, Suzuki, A, Yamane, T, Arai, M, Sakka, K, Omiya, K. | | Deposit date: | 2005-03-10 | | Release date: | 2005-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.52 Å) | | Cite: | Crystal Structure of Family 30 Carbohydrate Binding Module
To be Published
|
|
5B2A
 
 | | Crystal structure of a mutant of OspA | | Descriptor: | Outer surface protein A | | Authors: | Hori , Y, Makabe, K. | | Deposit date: | 2016-01-13 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Beta-sheet elasticity of peptide self-assembly mimic, PSAM, with a grafted sequence characterized by comprehensive analyses of isomorphous crystals
J Mol Liq, 290, 2019
|
|
6J48
 
 | | Glycine mutation on single layer beta-sheet of OspAsm1 | | Descriptor: | Outer surface protein A, TETRAETHYLENE GLYCOL | | Authors: | Makabe, K, Hori, Y. | | Deposit date: | 2019-01-08 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.201 Å) | | Cite: | Grafting a short chameleon sequence from alpha B crystallin into a beta-sheet scaffold protein.
Proteins, 87, 2019
|
|
6J47
 
 | |
6J49
 
 | | Grafting VLADV sequence into OspAsm1 | | Descriptor: | Outer surface protein A, TETRAETHYLENE GLYCOL | | Authors: | Makabe, K, Hori, Y. | | Deposit date: | 2019-01-08 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Grafting a short chameleon sequence from alpha B crystallin into a beta-sheet scaffold protein.
Proteins, 87, 2019
|
|
3GQB
 
 | | Crystal Structure of the A3B3 complex from V-ATPase | | Descriptor: | V-type ATP synthase alpha chain, V-type ATP synthase beta chain | | Authors: | Meher, M, Akimoto, S, Iwata, M, Nagata, K, Hori, Y, Yoshida, M, Yokoyama, S, Iwata, S, Yokoyama, K. | | Deposit date: | 2009-03-24 | | Release date: | 2009-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of A(3)B(3) complex of V-ATPase from Thermus thermophilus.
Embo J., 28, 2009
|
|
3AM2
 
 | | Clostridium perfringens enterotoxin | | Descriptor: | GLYCEROL, Heat-labile enterotoxin B chain, UNKNOWN ATOM OR ION | | Authors: | Kitadokoro, K, Nishimura, K, Kamitani, S, Kimura, J, Fukui, A, Abe, H, Horiguchi, Y. | | Deposit date: | 2010-08-12 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal Structure of Clostridium perfringens Enterotoxin Displays Features of {beta}-Pore-forming Toxins
J.Biol.Chem., 286, 2011
|
|
6LKR
 
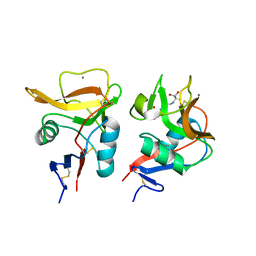 | | Crystal structure of mouse DCAR2 CRD domain complex | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, C-type lectin domain family 4, member b1, ... | | Authors: | Omahdi, Z, Horikawa, Y, Toyonaga, K, Kakuta, Y, Yamasaki, S. | | Deposit date: | 2019-12-20 | | Release date: | 2020-03-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural insight into the recognition of pathogen-derived phosphoglycolipids by C-type lectin receptor DCAR.
J.Biol.Chem., 295, 2020
|
|
6KZR
 
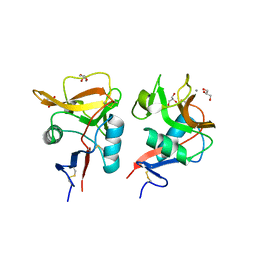 | | Crystal structure of mouse DCAR2 CRD domain | | Descriptor: | C-type lectin domain family 4, member b1, CALCIUM ION, ... | | Authors: | Omahdi, Z, Horikawa, Y, Toyonaga, K, Teramoto, T, Kakuta, Y, Yamasaki, S. | | Deposit date: | 2019-09-25 | | Release date: | 2020-03-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.304 Å) | | Cite: | Structural insight into the recognition of pathogen-derived phosphoglycolipids by C-type lectin receptor DCAR.
J.Biol.Chem., 295, 2020
|
|
6LFJ
 
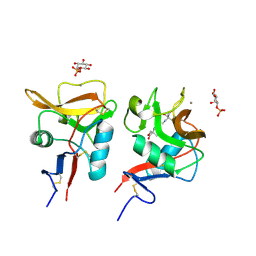 | | Crystal structure of mouse DCAR2 CRD domain complex with IPM2 | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, C-type lectin domain family 4, member b1, ... | | Authors: | Omahdi, Z, Horikawa, Y, Toyonaga, K, Kakuta, Y, Yamasaki, S. | | Deposit date: | 2019-12-03 | | Release date: | 2020-03-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural insight into the recognition of pathogen-derived phosphoglycolipids by C-type lectin receptor DCAR.
J.Biol.Chem., 295, 2020
|
|
1VE0
 
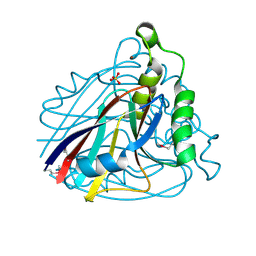 | | Crystal structure of uncharacterized protein ST2072 from Sulfolobus tokodaii | | Descriptor: | SULFATE ION, ZINC ION, hypothetical protein (ST2072) | | Authors: | Tanabe, E, Horiike, Y, Tsumoto, K, Yasutake, Y, Yao, M, Tanaka, I, Kumagai, I. | | Deposit date: | 2004-03-26 | | Release date: | 2005-03-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the uncharacterized protein ST2072 from Sulfolobus tokodaii
To be Published
|
|
2EC5
 
 | |
2EBF
 
 | |
2EBH
 
 | |
7X2B
 
 | | Red fluorescent protein from Diadumene lineata | | Descriptor: | Red fluorescent protein, SULFATE ION | | Authors: | Makabe, K, Hotta, J, Mizuno, H. | | Deposit date: | 2022-02-25 | | Release date: | 2023-03-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Cloning and structural basis of fluorescent protein color variants from identical species of sea anemone, Diadumene lineata.
Photochem Photobiol Sci, 22, 2023
|
|
5B71
 
 | | Crystal structure of complement C5 in complex with SKY59 | | Descriptor: | Complement C5 beta chain, SKY59 Fab heavy chain, SKY59 Fab light chain | | Authors: | Irie, M, Shimizu, Y, Sampei, Z, Fukuzawa, T. | | Deposit date: | 2016-06-03 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Long lasting neutralization of C5 by SKY59, a novel recycling antibody, is a potential therapy for complement-mediated diseases.
Sci Rep, 7, 2017
|
|
8W6X
 
 | | Neutron structure of [NiFe]-hydrogenase from D. vulgaris Miyazaki F in its oxidized state | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, FE3-S4 CLUSTER, ... | | Authors: | Hiromoto, T, Tamada, T. | | Deposit date: | 2023-08-30 | | Release date: | 2023-09-13 | | Last modified: | 2023-11-15 | | Method: | NEUTRON DIFFRACTION (1.04 Å), X-RAY DIFFRACTION | | Cite: | New insights into the oxidation process from neutron and X-ray crystal structures of an O 2 -sensitive [NiFe]-hydrogenase.
Chem Sci, 14, 2023
|
|
6M9L
 
 | | Structure-based Design, Synthesis, and Biological Evaluation of Imidazo[4,5-b]pyridine-2-one based p38 MAP Kinase Inhibitors by scaffold hopping - compound 10 | | Descriptor: | 3-benzyl-6-[(2,4-difluorophenyl)amino]-1,3-dihydro-2H-imidazo[4,5-b]pyridin-2-one, Mitogen-activated protein kinase 14 | | Authors: | Lane, W, Okada, K. | | Deposit date: | 2018-08-23 | | Release date: | 2019-04-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure-Based Design, Synthesis, and Biological Evaluation of Imidazo[4,5-b]pyridin-2-one-Based p38 MAP Kinase Inhibitors: Part 1.
Chemmedchem, 14, 2019
|
|
6M95
 
 | | Structure-based Design, Synthesis, and Biological Evaluation of Imidazo[4,5-b]pyridine-2-one based p38 MAP Kinase Inhibitors by scaffold hopping: compound 1 | | Descriptor: | (4-benzylpiperidin-1-yl)[2-methoxy-4-(methylsulfanyl)phenyl]methanone, Mitogen-activated protein kinase 14 | | Authors: | Lane, W, Okada, K. | | Deposit date: | 2018-08-22 | | Release date: | 2019-04-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Based Design, Synthesis, and Biological Evaluation of Imidazo[4,5-b]pyridin-2-one-Based p38 MAP Kinase Inhibitors: Part 1.
Chemmedchem, 14, 2019
|
|
5WJJ
 
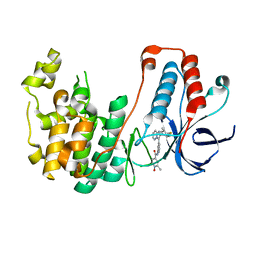 | | Structure-based Design, Synthesis, and Biological Evaluation of Imidazo[1,2-b]pyridazine-based p38 MAP Kinase Inhibitors | | Descriptor: | Mitogen-activated protein kinase 14, N-{4-[2-(4-fluoro-3-methylphenyl)imidazo[1,2-b]pyridazin-3-yl]pyridin-2-yl}-2-methyl-1-oxo-1lambda~5~-pyridine-4-carboxamide | | Authors: | Snell, G.P, Okada, K, Bragstad, K, Sang, B.-C. | | Deposit date: | 2017-07-23 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-based design, synthesis, and biological evaluation of imidazo[1,2-b]pyridazine-based p38 MAP kinase inhibitors.
Bioorg. Med. Chem., 26, 2018
|
|
6OHD
 
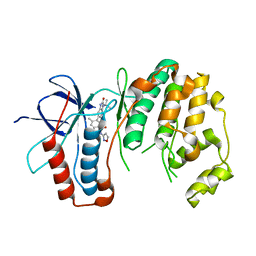 | | P38 in complex with T-3220137 | | Descriptor: | 3-(3-tert-butyl-2-oxo-2,3-dihydro-1H-imidazo[4,5-b]pyridin-6-yl)-4-methyl-N-(1,2-oxazol-3-yl)benzamide, Mitogen-activated protein kinase 14 | | Authors: | Lane, W, Saikatendu, K. | | Deposit date: | 2019-04-05 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Based Design, Synthesis, and Biological Evaluation of Imidazo[4,5-b]Pyridin-2-one-Based p38 MAP Kinase Inhibitors: Part 2.
Chemmedchem, 14, 2019
|
|
6ANL
 
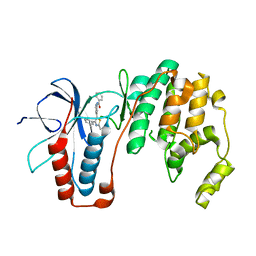 | | Structure-based Design, Synthesis, and Biological Evaluation of Imidazo[1,2-b]pyridazine-based p38 MAP Kinase Inhibitors | | Descriptor: | Mitogen-activated protein kinase 14, TAK-715 | | Authors: | Snell, G.P, Okada, K, Bragstad, K, Sang, B.-C. | | Deposit date: | 2017-08-14 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design, synthesis, and biological evaluation of imidazo[1,2-b]pyridazine-based p38 MAP kinase inhibitors.
Bioorg. Med. Chem., 26, 2018
|
|
6J6C
 
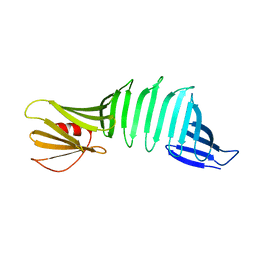 | |
