2FNF
 
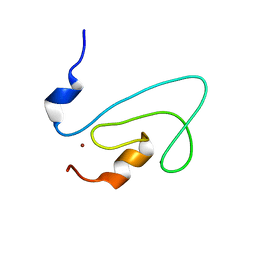 | | C1 domain of Nore1 | | Descriptor: | ZINC ION, putative Ras Effector Nore1 | | Authors: | Harjes, E, Harjes, S, Wohlgemuth, S, Krieger, E, Herrmann, C, Muller, K.H, Bayer, P. | | Deposit date: | 2006-01-11 | | Release date: | 2006-02-07 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | GTP-Ras disrupts the intramolecular complex of C1 and RA domains of Nore1.
Structure, 14, 2006
|
|
6U1L
 
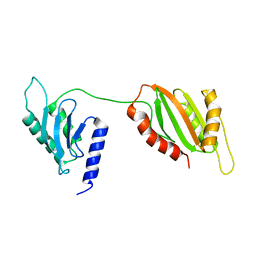 | | Structure of two-domain translational regulator Yih1 reveals a possible mechanism of action | | Descriptor: | Protein IMPACT homolog | | Authors: | Harjes, E, Jameson, G.B, Edwards, P.J.B, Goroncy, A.K, Loo, T, Norris, G.E. | | Deposit date: | 2019-08-16 | | Release date: | 2021-02-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Experimentally based structural model of Yih1 provides insight into its function in controlling the key translational regulator Gcn2.
Febs Lett., 595, 2021
|
|
6U1O
 
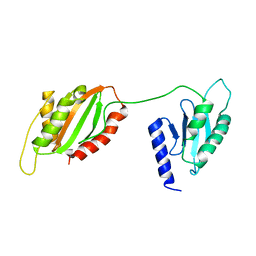 | | Structure of two-domain translational regulator Yih1 reveals a possible mechanism of action | | Descriptor: | Protein IMPACT homolog | | Authors: | Harjes, E, Jameson, G.B, Edwards, P.J.B, Goroncy, A.K, Loo, T, Norris, G.E. | | Deposit date: | 2019-08-16 | | Release date: | 2021-02-10 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Experimentally based structural model of Yih1 provides insight into its function in controlling the key translational regulator Gcn2.
Febs Lett., 595, 2021
|
|
6BQI
 
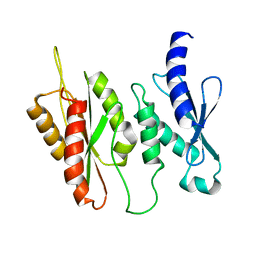 | | Structure of two-domain translational regulator Yih1 reveals a possible mechanism of action | | Descriptor: | Protein IMPACT homolog | | Authors: | Harjes, E, Jameson, G.B, Edwards, P.J.B, Goroncy, A.K, Loo, T, Norris, G.E. | | Deposit date: | 2017-11-27 | | Release date: | 2018-11-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Experimentally based structural model of Yih1 provides insight into its function in controlling the key translational regulator Gcn2.
Febs Lett., 2020
|
|
8DFZ
 
 | |
2KEM
 
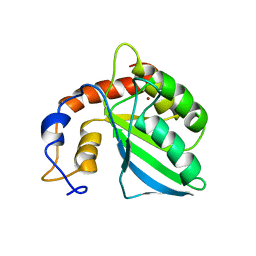 | | Extended structure of citidine deaminase domain of APOBEC3G | | Descriptor: | DNA dC->dU-editing enzyme APOBEC-3G, ZINC ION | | Authors: | Harjes, E, Gross, P.J, Chen, K, Lu, Y, Shindo, K, Nowarski, R, Gross, J.D, Kotler, M, Harris, R.S, Matsuo, H. | | Deposit date: | 2009-01-30 | | Release date: | 2009-06-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | An extended structure of the APOBEC3G catalytic domain suggests a unique holoenzyme model
J.Mol.Biol., 389, 2009
|
|
8FIK
 
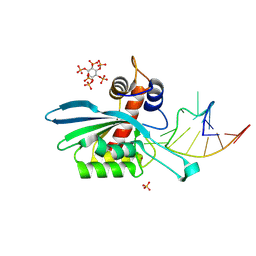 | | APOBEC3A E72A inactive mutant in complex with ATTC-hairpin DNA substrate | | Descriptor: | CHLORIDE ION, DNA (5'-D(P*CP*CP*CP*AP*TP*CP*AP*TP*TP*CP*GP*AP*TP*GP*GP*G)-3'), DNA dC->dU-editing enzyme APOBEC-3A, ... | | Authors: | Harjes, S, Jameson, G.B, Harjes, E, Filichev, V.V, Kurup, H.M. | | Deposit date: | 2022-12-16 | | Release date: | 2023-09-06 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.912 Å) | | Cite: | Structure-guided inhibition of the cancer DNA-mutating enzyme APOBEC3A.
Nat Commun, 14, 2023
|
|
8FIL
 
 | | Zinc-free APOBEC3A (inactive E72A mutant) in complex with TTC-hairpin DNA substrate | | Descriptor: | CHLORIDE ION, DNA (5'-D(*TP*GP*CP*GP*CP*TP*TP*CP*GP*CP*GP*CP*T)-3'), DNA dC->dU-editing enzyme APOBEC-3A, ... | | Authors: | Harjes, S, Jameson, G.B, Harjes, E, Filichev, V.V, Kurup, H.M. | | Deposit date: | 2022-12-16 | | Release date: | 2023-09-06 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure-guided inhibition of the cancer DNA-mutating enzyme APOBEC3A.
Nat Commun, 14, 2023
|
|
8FII
 
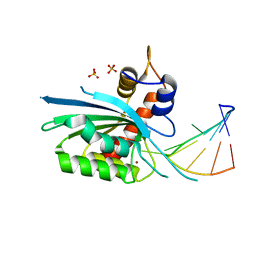 | | Wild type APOBEC3A in complex with TT(FdZ)-hairpin inhibitor (crystal form 1) | | Descriptor: | DNA (5'-D(P*GP*CP*GP*CP*TP*TP*(UFP)P*GP*CP*GP*C)-3'), DNA dC->dU-editing enzyme APOBEC-3A, PHOSPHATE ION, ... | | Authors: | Harjes, S, Jameson, G.B, Harjes, E, Filichev, V.V, Kurup, H.M. | | Deposit date: | 2022-12-16 | | Release date: | 2023-09-06 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Structure-guided inhibition of the cancer DNA-mutating enzyme APOBEC3A.
Nat Commun, 14, 2023
|
|
8FIJ
 
 | | Wild type APOBEC3A in complex with TT(FdZ)-hairpin inhibitor (crystal form 2) | | Descriptor: | DNA (5'-D(*GP*CP*GP*CP*TP*TP*(UFP)P*GP*CP*GP*CP*T)-3'), DNA dC->dU-editing enzyme APOBEC-3A, PHOSPHATE ION, ... | | Authors: | Harjes, S, Jameson, G.B, Harjes, E, Filichev, V.V, Kurup, H.M. | | Deposit date: | 2022-12-16 | | Release date: | 2023-09-06 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Structure-guided inhibition of the cancer DNA-mutating enzyme APOBEC3A.
Nat Commun, 14, 2023
|
|
8FIM
 
 | | Structure of APOBEC3A (E72A inactive mutant) in complex with TTC-hairpin DNA substrate | | Descriptor: | CHLORIDE ION, DNA (5'-D(*TP*GP*CP*GP*CP*TP*TP*CP*GP*CP*GP*CP*T)-3'), DNA dC->dU-editing enzyme APOBEC-3A, ... | | Authors: | Harjes, S, Jameson, G.B, Harjes, E, Filichev, V.V, Kurup, H.M. | | Deposit date: | 2022-12-16 | | Release date: | 2023-09-06 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structure-guided inhibition of the cancer DNA-mutating enzyme APOBEC3A.
Nat Commun, 14, 2023
|
|
2JYW
 
 | | Solution structure of C-terminal domain of APOBEC3G | | Descriptor: | DNA dC->dU-editing enzyme APOBEC-3G, ZINC ION | | Authors: | Chen, K, Harjes, E, Gross, P.J, Fahmy, A, Lu, Y, Shindo, K, Harris, R.S, Matsuo, H. | | Deposit date: | 2007-12-20 | | Release date: | 2008-02-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the DNA deaminase domain of the HIV-1 restriction factor APOBEC3G.
Nature, 452, 2008
|
|
6B34
 
 | |
6B35
 
 | |
2LQZ
 
 | |
3IR2
 
 | |
6ANM
 
 | |
6ANN
 
 | |
