6QNA
 
 | | Structure of bovine anti-RSV hybrid Fab B13HC-B4LC | | Descriptor: | B13 Heavy chain, B4 light chain, GLYCEROL | | Authors: | Ren, J, Nettleship, J.E, Harris, G, Mwangi, W, Rhaman, N, Grant, C, Kotecha, A, Fry, E, Charleston, B, Stuart, D.I, Hammond, J, Owens, R.J. | | Deposit date: | 2019-02-10 | | Release date: | 2019-05-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | The role of the light chain in the structure and binding activity of two cattle antibodies that neutralize bovine respiratory syncytial virus.
Mol.Immunol., 112, 2019
|
|
6QN9
 
 | | Structure of bovine anti-RSV Fab B4 | | Descriptor: | GLYCEROL, Heavy chain, SULFATE ION, ... | | Authors: | Ren, J, Nettleship, J.E, Harris, G, Mwangi, W, Rhaman, N, Grant, C, Kotecha, A, Fry, E, Charleston, B, Stuart, D.I, Hammond, J, Owens, R.J. | | Deposit date: | 2019-02-10 | | Release date: | 2019-05-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | The role of the light chain in the structure and binding activity of two cattle antibodies that neutralize bovine respiratory syncytial virus.
Mol.Immunol., 112, 2019
|
|
6QN7
 
 | | Structure of bovine anti-RSV hybrid Fab B4HC-B13LC | | Descriptor: | Heavy chain of bovine anti-RSV B4, Light chain of bovine anti-RSV B13 | | Authors: | Ren, J, Nettleship, J.E, Harris, G, Mwangi, W, Rhaman, N, Grant, C, Kotecha, A, Fry, E, Charleston, B, Stuart, D.I, Hammond, J, Owens, R.J. | | Deposit date: | 2019-02-10 | | Release date: | 2019-05-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The role of the light chain in the structure and binding activity of two cattle antibodies that neutralize bovine respiratory syncytial virus.
Mol.Immunol., 112, 2019
|
|
6QN8
 
 | | Structure of bovine anti-RSV Fab B13 | | Descriptor: | CHLORIDE ION, Heavy chain of bovine anti-RSV B13 Fab, Light chain of bovine anti-RSV Fab B13 | | Authors: | Ren, J, Nettleship, J.E, Harris, G, Mwangi, W, Rhaman, N, Grant, C, Kotecha, A, Fry, E, Charleston, B, Stuart, D.I, Hammond, J, Owens, R.J. | | Deposit date: | 2019-02-10 | | Release date: | 2019-05-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | The role of the light chain in the structure and binding activity of two cattle antibodies that neutralize bovine respiratory syncytial virus.
Mol.Immunol., 112, 2019
|
|
3TTE
 
 | | Crystal structure of enolase brado_4202 (target EFI-501651) from Bradyrhizobium complexed with magnesium and mandelic acid | | Descriptor: | (S)-MANDELIC ACID, FORMIC ACID, GLYCEROL, ... | | Authors: | Patskovsky, Y, Kim, J, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammond, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-09-14 | | Release date: | 2011-09-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Mandelate Racemase from Bradyrhizobium Sp. Ors278
To be Published
|
|
3TOY
 
 | | CRYSTAL STRUCTURE OF ENOLASE BRADO_4202 (TARGET EFI-501651) FROM Bradyrhizobium sp. ORS278 WITH CALCIUM AND ACETATE BOUND | | Descriptor: | ACETATE ION, CALCIUM ION, Mandelate racemase/muconate lactonizing enzyme family protein, ... | | Authors: | Patskovsky, Y, Kim, J, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammond, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-09-06 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | CRYSTAL STRUCTURE OF MANDELATE RACEMASE FROM Bradyrhizobium sp. ORS278
To be Published
|
|
4O9K
 
 | | Crystal structure of the CBS pair of a putative D-arabinose 5-phosphate isomerase from Methylococcus capsulatus in complex with CMP-Kdo | | Descriptor: | Arabinose 5-phosphate isomerase, CYTIDINE 5'-MONOPHOSPHATE 3-DEOXY-BETA-D-GULO-OCT-2-ULO-PYRANOSONIC ACID, GLYCEROL | | Authors: | Shabalin, I.G, Cooper, D.R, Shumilin, I.A, Zimmerman, M.D, Majorek, K.A, Hammonds, J, Hillerich, B.S, Nawar, A, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-01-02 | | Release date: | 2014-01-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure and kinetic properties of D-arabinose 5-phosphate isomerase from Methylococcus capsulatus
To be Published
|
|
4UC0
 
 | | Crystal Structure Of a purine nucleoside phosphorylase (PSI-NYSGRC-029736) from Agrobacterium vitis | | Descriptor: | HYPOXANTHINE, Purine nucleoside phosphorylase | | Authors: | Cameron, S.A, Sampathkumar, P, Ramagopal, U.A, Attonito, J, Ahmed, M, Bhosle, R, Bonanno, J, Chamala, S, Chowdhury, S, Glenn, A.S, Hammonds, J, Hillerich, B, Love, J.D, Seidel, R, Stead, M, Toro, R, Wasserman, S.R, Schramm, V.L, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-08-13 | | Release date: | 2014-10-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure Of a purine nucleoside phosphorylase (PSI-NYSGRC-029736) from Agrobacterium vitis
To be published
|
|
4WCZ
 
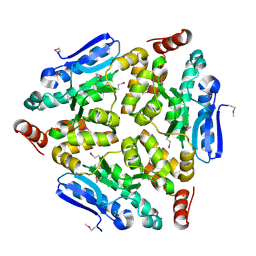 | | Crystal structure of a putative enoyl-CoA hydratase/isomerase from Novosphingobium aromaticivorans | | Descriptor: | Enoyl-CoA hydratase/isomerase | | Authors: | Tkaczuk, K.L, Cooper, D.R, Chapman, H.C, Niedzialkowska, E, Cymborowski, M.T, Hillerich, B.S, Stead, M, Ahmed, M, Hammonds, J, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-09-05 | | Release date: | 2014-12-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of a putative enoyl-CoA hydratase/isomerase from Novosphingobium aromaticivorans
to be published
|
|
3UF0
 
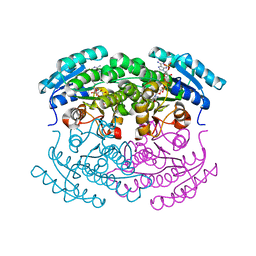 | | Crystal structure of a putative NAD(P) dependent gluconate 5-dehydrogenase from Beutenbergia cavernae(EFI target EFI-502044) with bound NADP (low occupancy) | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short-chain dehydrogenase/reductase SDR | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Hillerich, B, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-10-31 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a putative NAD(P) dependent gluconate 5-dehydrogenase from Beutenbergia cavernae(EFI target EFI-502044) with bound NADP (low occupancy)
To be Published
|
|
4U13
 
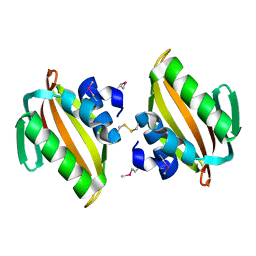 | | Crystal structure of putative polyketide cyclase (protein SMa1630) from Sinorhizobium meliloti at 2.3 A resolution | | Descriptor: | putative polyketide cyclase SMa1630 | | Authors: | Shabalin, I.G, Bacal, P, Osinski, T, Cooper, D.R, Szlachta, K, Stead, M, Grabowski, M, Hammonds, J, Ahmed, M, Hillerich, B.S, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-07-14 | | Release date: | 2014-09-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a putative polyketide cyclase (protein SMa1630) from Sinorhizobium meliloti at 2.3 A resolution
to be published
|
|
4G9H
 
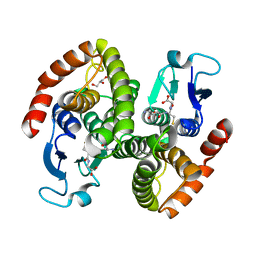 | | Crystal structure of glutahtione s-transferase homolog from yersinia pestis, target EFI-501894, with bound glutathione | | Descriptor: | GLUTATHIONE, GLYCEROL, Glutathione S-transferase | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-07-23 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of glutahtione s-transferase homolog from yersinia pestis, target efi-501894, with bound glutathione
To be Published
|
|
4GF0
 
 | | Crystal structure of glutahtione transferase homolog from sulfitobacter, TARGET EFI-501084, with bound glutathione | | Descriptor: | CHLORIDE ION, GLUTATHIONE, Glutathione S-transferase | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-08-02 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of glutahtione transferase homolog from sulfitobacter, TARGET EFI-501084, with bound glutathione
To be Published
|
|
4GGH
 
 | | Crystal structure of an enolase family member from vibrio harveyi (efi-target 501692) with homology to mannonate dehydratase, with mg, hepes, and ethylene glycol bound (ordered loops, space group c2221) | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-08-06 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of an enolase family member from vibrio harveyi (efi-target 501692) with homology to mannonate dehydratase, with mg, hepes, and ethylene glycol bound (ordered loops, space group c2221)
To be Published
|
|
4FI4
 
 | | Crystal structure of mannonate dehydratase PRK15072 (TARGET EFI-502214) from Caulobacter sp. K31 | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-06-07 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Enolase Prk15072 (Target Efi-502214) from Caulobacter Sp. K31
To be Published
|
|
5C9G
 
 | | Crystal Structure of a Putative enoyl-CoA hydratase/isomerase family protein from Hyphomonas neptunium | | Descriptor: | D-MALATE, Enoyl-CoA hydratase/isomerase family protein, TETRAETHYLENE GLYCOL | | Authors: | Szlachta, K, Cooper, D.R, Chapman, H.C, Cymbrowski, M.T, Stead, M, Hillerich, B.S, Ahmed, M, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, Hammonds, J, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-06-26 | | Release date: | 2015-10-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of a Putative enoyl-CoA hydratase/isomerase family protein from Hyphomonas neptunium
to be published
|
|
2M4M
 
 | | Solution structure of the RRM domain of the hypothetical protein CAGL0M09691g from Candida glabrata | | Descriptor: | hypothetical protein | | Authors: | Harris, R, Hillerich, B, Ahmed, M, Bonanno, J.B, Chamala, S, Evans, B, Lafleur, J, Hammonds, J, Washington, E, Stead, M, Love, J, Attonito, J, Patel, H, Seidel, R.D, Chook, Y.M, Rout, M.P, Girvin, M.E, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Nucleocytoplasmic Transport: a Target for Cellular Control (NPCXstals) | | Deposit date: | 2013-02-07 | | Release date: | 2013-02-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RRM domain of the hypothetical protein CAGL0M09691g from Candida glabrata
To be Published
|
|
2M4N
 
 | | Solution structure of the putative Ras interaction domain of AFD-1, isoform a from Caenorhabditis elegans | | Descriptor: | Protein AFD-1, isoform a | | Authors: | Harris, R, Hillerich, B, Ahmed, M, Bonanno, J.B, Chamala, S, Evans, B, Lafleur, J, Hammonds, J, Washington, E, Stead, M, Love, J, Attonito, J, Seidel, R.D, Liddington, R.C, Weis, W.I, Nelson, W.J, Girvin, M.E, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Assembly, Dynamics and Evolution of Cell-Cell and Cell-Matrix Adhesions (CELLMAT) | | Deposit date: | 2013-02-07 | | Release date: | 2013-03-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the putative Ras interaction domain of AFD-1, isoform a from Caenorhabditis elegans
To be Published
|
|
4Q60
 
 | | Crystal structure of a 4-hydroxyproline epimerase from Burkholderia Multivorans atcc 17616, target EFI-506586, open form, with bound pyrrole-2-carboxylate | | Descriptor: | GLYCEROL, PROLINE RACEMASE, PYRROLE-2-CARBOXYLATE | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Sojitra, S, Stead, M, Washington, E, Glenn, A.S, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-04-18 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | CRYSTAL STRUCTURE OF PROLINE RACEMASE Bmul_4447 FROM Burkholderia multivorans, TARGET EFI-506586
To be Published
|
|
4PX1
 
 | | CRYSTAL STRUCTURE OF Maleylacetoacetate isomerase from Methylobacteriu extorquens AM1 WITH BOUND MALONATE (TARGET EFI-507068) | | Descriptor: | CHLORIDE ION, MALONIC ACID, Maleylacetoacetate isomerase (Glutathione S-transferase) | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-03-21 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of glutathione s-transferase zeta from methylobacterium extorquens (TARGET EFI-507068)
To be Published
|
|
4PXO
 
 | | Crystal structure of Maleylacetoacetate isomerase from Methylobacteriu extorquens AM1 WITH BOUND MALONATE AND GSH (TARGET EFI-507068) | | Descriptor: | 1,2-ETHANEDIOL, GLUTATHIONE, MALONIC ACID, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-03-24 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of glutathione s-transferase zeta from Methylobacterium extorquens (TARGET EFI-507068)
To be Published
|
|
4Q2H
 
 | | Crystal structure of probable proline racemase from agrobacterium radiobacter K84, TARGET EFI-506561, with bound carbonate | | Descriptor: | BICARBONATE ION, GLYCEROL, Proline racemase protein | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Washington, E, Glenn, A.S, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-04-08 | | Release date: | 2014-04-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Proline Racemase Arad_0731 from Agrobacterium Radiobacter, Target Efi-506561
To be Published
|
|
4QRZ
 
 | | Crystal structure of sugar transporter atu4361 from agrobacterium fabrum c58, target efi-510558, with bound maltotriose | | Descriptor: | ABC-TYPE SUGAR TRANSPORTER, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Stead, M, Washington, E, Glenn, A.S, Chowdhury, S, Evans, B, Hammonds, J, Love, J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-07-02 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Crystal Structure of Maltoside Transporter from Agrobacterium Radiobacter, Target Efi-510558
To be Published
|
|
2M72
 
 | | Solution structure of uncharacterized thioredoxin-like protein PG_2175 from Porphyromonas gingivalis | | Descriptor: | Uncharacterized thioredoxin-like protein | | Authors: | Harris, R, Ahmed, M, Attonito, J, Bonanno, J.B, Chamala, S, Chowdhury, S, Evans, B, Fiser, A, Glenn, A.S, Hammonds, J, Hillerich, B, Khafizov, K, Lafleur, J, Love, J.D, Seidel, R.D, Stead, M, Girvin, M.E, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-16 | | Release date: | 2013-05-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of uncharacterized thioredoxin-like protein PG_2175 from Porphyromonas gingivalis
To be Published
|
|
2M71
 
 | | Solution structure of the a C-terminal domain of translation initiation factor IF-3 from Campylobacter jejuni | | Descriptor: | Translation initiation factor IF-3 | | Authors: | Harris, R, Ahmed, M, Attonito, J, Bonanno, J.B, Chamala, S, Chowdhury, S, Evans, B, Fiser, A, Glenn, A.S, Hammonds, J, Hillerich, B, Khafizov, K, Lafleur, J, Love, J.D, Seidel, R.D, Stead, M, Girvin, M.E, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-16 | | Release date: | 2013-05-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the a C-terminal domain of translation initiation factor IF-3 from Campylobacter jejuni
To be Published
|
|
