8CD9
 
 | | Cathepsin B1 from Schistosoma mansoni in complex with gallinamide analog 6 | | Descriptor: | 1,2-ETHANEDIOL, Cathepsin B-like peptidase (C01 family), SODIUM ION, ... | | Authors: | Rubesova, P, Brynda, J, Fanfrlik, J, Gerwick, W.H, Mares, M. | | Deposit date: | 2023-01-30 | | Release date: | 2024-02-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Nature-Inspired Gallinamides Are Potent Antischistosomal Agents: Inhibition of the Cathepsin B1 Protease Target and Binding Mode Analysis.
Acs Infect Dis., 10, 2024
|
|
7JUJ
 
 | | Cruzain bound to Gallinamide inhibitor | | Descriptor: | Cruzipain, POTASSIUM ION, gallinamide A, ... | | Authors: | Silva, E.B, Sharma, V, Alvarez, L.H, Gerwick, W.H, McKerrow, J.H, Podust, L.M. | | Deposit date: | 2020-08-19 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Intramolecular Interactions Enhance the Potency of Gallinamide A Analogues against Trypanosoma cruzi .
J.Med.Chem., 65, 2022
|
|
8CC2
 
 | | Cathepsin B1 from Schistosoma mansoni in complex with gallinamide A | | Descriptor: | Cathepsin B-like peptidase (C01 family), SODIUM ION, gallinamide A, ... | | Authors: | Rubesova, P, Brynda, J, Mares, M, Gerwick, W.H. | | Deposit date: | 2023-01-26 | | Release date: | 2024-02-07 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Nature-Inspired Gallinamides Are Potent Antischistosomal Agents: Inhibition of the Cathepsin B1 Protease Target and Binding Mode Analysis.
Acs Infect Dis., 10, 2024
|
|
8CCU
 
 | | Cathepsin B1 from Schistosoma mansoni in complex with gallinamide analog 1 | | Descriptor: | Cathepsin B-like peptidase (C01 family), SODIUM ION, [(2~{S})-1-[[(2~{S})-1-[[(2~{S})-5-[(2~{S})-3-methoxy-2-(2-methylpropyl)-5-oxidanylidene-2~{H}-pyrrol-1-yl]-5-oxidanylidene-pentan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl] (2~{S},3~{S})-2-(dimethylamino)-3-methyl-pentanoate | | Authors: | Rubesova, P, Brynda, J, Gerwick, W.H, Mares, M. | | Deposit date: | 2023-01-27 | | Release date: | 2024-02-07 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Nature-Inspired Gallinamides Are Potent Antischistosomal Agents: Inhibition of the Cathepsin B1 Protease Target and Binding Mode Analysis.
Acs Infect Dis., 10, 2024
|
|
5KP7
 
 | |
5KP6
 
 | |
5KP8
 
 | |
5KP5
 
 | |
6B3A
 
 | |
6B3B
 
 | | AprA Methyltransferase 1 - GNAT in complex with Mn2+ , SAM, and Malonate | | Descriptor: | AprA Methyltransferase 1, GLYCEROL, MALONATE ION, ... | | Authors: | Skiba, M.A, Smith, J.L. | | Deposit date: | 2017-09-21 | | Release date: | 2017-11-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Mononuclear Iron-Dependent Methyltransferase Catalyzes Initial Steps in Assembly of the Apratoxin A Polyketide Starter Unit.
ACS Chem. Biol., 12, 2017
|
|
6B39
 
 | | AprA Methyltransferase 1 - GNAT in complex with SAH | | Descriptor: | AprA Methyltransferase 1, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION | | Authors: | Skiba, M.A, Smith, J.L. | | Deposit date: | 2017-09-21 | | Release date: | 2017-11-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.392 Å) | | Cite: | A Mononuclear Iron-Dependent Methyltransferase Catalyzes Initial Steps in Assembly of the Apratoxin A Polyketide Starter Unit.
ACS Chem. Biol., 12, 2017
|
|
6D6Y
 
 | |
3U1T
 
 | | Haloalkane Dehalogenase, DmmA, of marine microbial origin | | Descriptor: | CHLORIDE ION, DmmA Haloalkane Dehalogenase, MALONATE ION | | Authors: | Gehret, J.J, Smith, J.L. | | Deposit date: | 2011-09-30 | | Release date: | 2011-12-28 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and activity of DmmA, a marine haloalkane dehalogenase.
Protein Sci., 21, 2012
|
|
7UCH
 
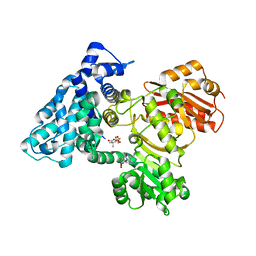 | | AprA Methyltransferase 1 - GNAT in complex with Mn2+ , SAM, and Di-methyl-malonate | | Descriptor: | AprA Methyltransferase 1, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Skiba, M.A, Lao, Y, Smith, J.L. | | Deposit date: | 2022-03-16 | | Release date: | 2022-06-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural Basis for Control of Methylation Extent in Polyketide Synthase Metal-Dependent C -Methyltransferases.
Acs Chem.Biol., 17, 2022
|
|
4GOX
 
 | |
4GBM
 
 | |
2REE
 
 | |
6TH7
 
 | |
3QIT
 
 | |
5THY
 
 | |
5THZ
 
 | |
6MFD
 
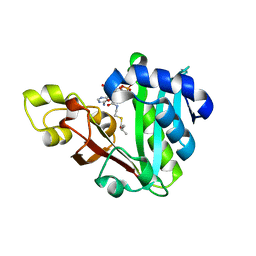 | | GphF GNAT-like decarboxylase in complex with isobutyryl-CoA | | Descriptor: | ACETATE ION, GLYCEROL, GphF, ... | | Authors: | Skiba, M.A, Tran, C.L, Smith, J.L. | | Deposit date: | 2018-09-10 | | Release date: | 2019-09-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.794 Å) | | Cite: | Repurposing the GNAT Fold in the Initiation of Polyketide Biosynthesis.
Structure, 28, 2020
|
|
6MFC
 
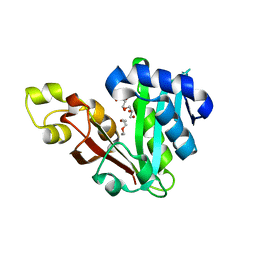 | | GphF GNAT-like decarboxylase | | Descriptor: | GLYCEROL, GphF, PENTAETHYLENE GLYCOL | | Authors: | Skiba, M.A, Tran, C.L, Smith, J.L. | | Deposit date: | 2018-09-10 | | Release date: | 2019-09-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.589 Å) | | Cite: | Repurposing the GNAT Fold in the Initiation of Polyketide Biosynthesis.
Structure, 28, 2020
|
|
6ECT
 
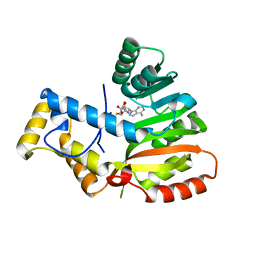 | | StiE O-MT residues 961-1257 | | Descriptor: | S-ADENOSYLMETHIONINE, StiE protein | | Authors: | Skiba, M.A, Bivins, M.M, Smith, J.L. | | Deposit date: | 2018-08-08 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structural Basis of Polyketide Synthase O-Methylation.
ACS Chem. Biol., 13, 2018
|
|
6ECU
 
 | |
