1QKS
 
 | | CYTOCHROME CD1 NITRITE REDUCTASE, OXIDISED FORM | | Descriptor: | CYTOCHROME CD1 NITRITE REDUCTASE, GLYCEROL, HEME C, ... | | Authors: | Fulop, V. | | Deposit date: | 1999-08-05 | | Release date: | 1999-08-18 | | Last modified: | 2019-10-09 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | The anatomy of a bifunctional enzyme: structural basis for reduction of oxygen to water and synthesis of nitric oxide by cytochrome cd1.
Cell(Cambridge,Mass.), 81, 1995
|
|
1E8N
 
 | | PROLYL OLIGOPEPTIDASE FROM PORCINE BRAIN, MUTANT, COMPLEXED WITH PEPTIDE | | Descriptor: | GLYCEROL, PEPTIDE INHIBITOR, PROLYL ENDOPEPTIDASE | | Authors: | Fulop, V. | | Deposit date: | 2000-09-27 | | Release date: | 2001-01-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of Prolyl Oligopeptidase Substrate/ Inhibitor Complexes. Use of Inhibitor Binding for Titration of the Catalytic Histidine Residue
J.Biol.Chem., 276, 2001
|
|
1E8M
 
 | | PROLYL OLIGOPEPTIDASE FROM PORCINE BRAIN, MUTANT, COMPLEXED WITH INHIBITOR | | Descriptor: | GLYCEROL, N-[(benzyloxy)carbonyl]glycyl-L-proline, PROLYL ENDOPEPTIDASE | | Authors: | Fulop, V. | | Deposit date: | 2000-09-27 | | Release date: | 2001-01-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of Prolyl Oligopeptidase Substrate/ Inhibitor Complexes. Use of Inhibitor Binding for Titration of the Catalytic Histidine Residue
J.Biol.Chem., 276, 2001
|
|
1QFS
 
 | |
2YK3
 
 | | CRITHIDIA FASCICULATA CYTOCHROME C | | Descriptor: | CYTOCHROME C, HEME C, SULFATE ION | | Authors: | Fulop, V, Sam, K.A, Ferguson, S.J, Ginger, M.L, Allen, J.W.A. | | Deposit date: | 2011-05-25 | | Release date: | 2011-06-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of a Trypanosomatid Mitochondrial Cytochrome C with Heme Attached Via Only One Thioether Bond and Implications for the Substrate Recognition Requirements of Heme Lyase.
FEBS J., 276, 2009
|
|
1E5T
 
 | | PROLYL OLIGOPEPTIDASE FROM PORCINE BRAIN, MUTANT | | Descriptor: | GLYCEROL, PROLYL ENDOPEPTIDASE | | Authors: | Fulop, V. | | Deposit date: | 2000-08-02 | | Release date: | 2000-10-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Catalysis of Serine Oligopeptidases is Controlled by a Gating Filter Mechanism
Embo Rep., 1, 2000
|
|
1QFM
 
 | | PROLYL OLIGOPEPTIDASE FROM PORCINE MUSCLE | | Descriptor: | 1-HYDROXY-1-THIO-GLYCEROL, GLYCEROL, MONOTHIOGLYCEROL, ... | | Authors: | Fulop, V. | | Deposit date: | 1999-04-12 | | Release date: | 1999-05-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Prolyl oligopeptidase: an unusual beta-propeller domain regulates proteolysis.
Cell(Cambridge,Mass.), 94, 1998
|
|
1EB7
 
 | |
1EBE
 
 | | Laue diffraction study on the structure of cytochrome c peroxidase compound I | | Descriptor: | CYTOCHROME C PEROXIDASE, OXYGEN ATOM, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Fulop, V, Phizackerley, R.P, Soltis, S.M, Clifton, I.J, Wakatsuki, S, Erman, J.E, Hajdu, J, Edwards, S.L. | | Deposit date: | 2001-07-25 | | Release date: | 2001-07-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Laue Diffraction Study on the Structure of Cytochrome C Peroxidase Compound I
Structure, 2, 1994
|
|
5OLJ
 
 | | Crystal structure of Porphyromonas gingivalis dipeptidyl peptidase 4 | | Descriptor: | Dipeptidyl peptidase IV, GLYCEROL | | Authors: | Fulop, V. | | Deposit date: | 2017-07-27 | | Release date: | 2017-09-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Porphyromonas gingivalis dipeptidyl peptidase 4 and structure-activity relationships based on inhibitor profiling.
Eur J Med Chem, 139, 2017
|
|
1H2X
 
 | | PROLYL OLIGOPEPTIDASE FROM PORCINE BRAIN, Y473F MUTANT | | Descriptor: | GLYCEROL, PROLYL ENDOPEPTIDASE | | Authors: | Fulop, V. | | Deposit date: | 2002-08-20 | | Release date: | 2002-11-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Electrostatic Effects and Binding Determinants in the Catalysis of Prolyl Oligopeptidase: Site Specific Mutagenesis at the Oxyanion Binding Site
J.Biol.Chem., 277, 2002
|
|
1H2W
 
 | | PROLYL OLIGOPEPTIDASE FROM PORCINE BRAIN | | Descriptor: | GLYCEROL, PROLYL ENDOPEPTIDASE | | Authors: | Fulop, V. | | Deposit date: | 2002-08-20 | | Release date: | 2002-11-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Electrostatic Effects and Binding Determinants in the Catalysis of Prolyl Oligopeptidase: Site Specific Mutagenesis at the Oxyanion Binding Site
J.Biol.Chem., 277, 2002
|
|
1E2R
 
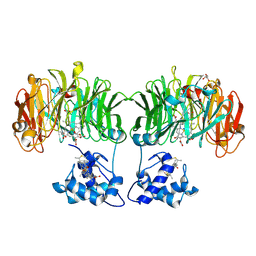 | | CYTOCHROME CD1 NITRITE REDUCTASE, REDUCED AND CYANIDE BOUND | | Descriptor: | CYANIDE ION, GLYCEROL, HEME C, ... | | Authors: | Fulop, V. | | Deposit date: | 2000-05-24 | | Release date: | 2000-05-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | X-Ray Crystallographic Study of Cyanide Binding Provides Insights Into the Structure-Function Relationship for Cytochrome Cd1 Nitrite Reductase from Paracoccus Pantotrophus.
J.Biol.Chem., 275, 2000
|
|
7NE9
 
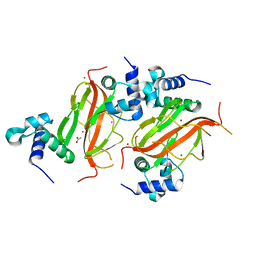 | |
6SRN
 
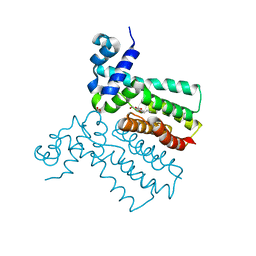 | |
6SNR
 
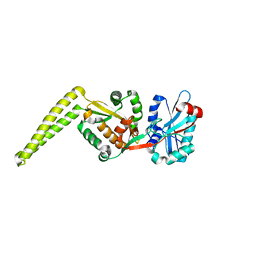 | | Crystal structure of FemX | | Descriptor: | Lipid II:glycine glycyltransferase | | Authors: | Fulop, V, Hinxman, K. | | Deposit date: | 2019-08-27 | | Release date: | 2020-09-09 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure-based modeling and dynamics of MurM, a Streptococcus pneumoniae penicillin resistance determinant present at the cytoplasmic membrane.
Structure, 29, 2021
|
|
6TJ2
 
 | |
6F0C
 
 | |
6F0B
 
 | |
6EVG
 
 | |
1GJM
 
 | |
4C12
 
 | | X-ray Crystal Structure of Staphylococcus aureus MurE with UDP-MurNAc- Ala-Glu-Lys and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Fulop, V, Roper, D.I, Ruane, K.M, Barreteau, H, Boniface, A, Dementin, S, Blanot, D, Mengin-Lecreulx, D, Gobec, S, Dessen, A, Dowson, C.G, Lloyd, A.J. | | Deposit date: | 2013-08-09 | | Release date: | 2013-10-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Specificity Determinants for Lysine Incorporation in Staphylococcus Aureus Peptidoglycan as Revealed by the Structure of a Mure Enzyme Ternary Complex.
J.Biol.Chem., 288, 2013
|
|
1VZ2
 
 | |
1VZ3
 
 | | PROLYL OLIGOPEPTIDASE FROM PORCINE BRAIN, T597C MUTANT | | Descriptor: | GLYCEROL, PROLYL ENDOPEPTIDASE | | Authors: | Rea, D, Fulop, V. | | Deposit date: | 2004-05-14 | | Release date: | 2004-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Concerted Structural Changes in the Peptidase and the Propeller Domains of Prolyl Oligopeptidase are Required for Substrate Binding
J.Mol.Biol., 340, 2004
|
|
1UOP
 
 | |
