5UE5
 
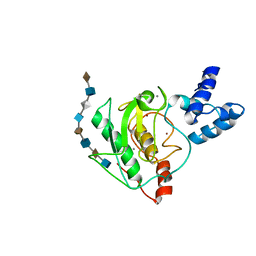 | | proMMP-7 with heparin octasaccharide bound to the catalytic domain | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, CALCIUM ION, Matrilysin, ... | | Authors: | Fulcher, Y.G, Prior, S.H, Linhardt, R.J, Van Doren, S.R. | | Deposit date: | 2016-12-29 | | Release date: | 2017-07-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Glycan Activation of a Sheddase: Electrostatic Recognition between Heparin and proMMP-7.
Structure, 25, 2017
|
|
5UE2
 
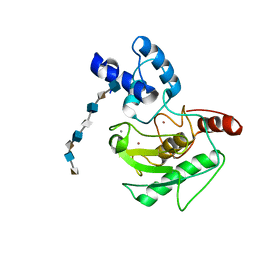 | | proMMP-7 with heparin octasaccharide bridging between domains | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, CALCIUM ION, Matrilysin, ... | | Authors: | Fulcher, Y.G, Prior, S.H, Linhardt, R.J, Van Doren, S.R. | | Deposit date: | 2016-12-29 | | Release date: | 2017-07-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Glycan Activation of a Sheddase: Electrostatic Recognition between Heparin and proMMP-7.
Structure, 25, 2017
|
|
7MY8
 
 | |
2MZE
 
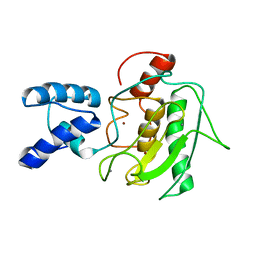 | | NMR Solution Structure of the PRO Form of Human Matrilysin (proMMP-7) | | Descriptor: | CALCIUM ION, Matrilysin, ZINC ION | | Authors: | Prior, S.H, Fulcher, Y.G, Van Doren, S.R. | | Deposit date: | 2015-02-11 | | Release date: | 2015-11-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Charge-Triggered Membrane Insertion of Matrix Metalloproteinase-7, Supporter of Innate Immunity and Tumors.
Structure, 23, 2015
|
|
2MLR
 
 | | Membrane Bilayer complex with Matrix Metalloproteinase-12 at its Alpha-face | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, Macrophage metalloelastase, ... | | Authors: | Koppisetti, R.K, Fulcher, Y.G, Prior, S.H, Lenoir, M, Overduin, M, Van Doren, S.R. | | Deposit date: | 2014-03-04 | | Release date: | 2014-12-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Ambidextrous binding of cell and membrane bilayers by soluble matrix metalloproteinase-12.
Nat Commun, 5, 2014
|
|
2MLS
 
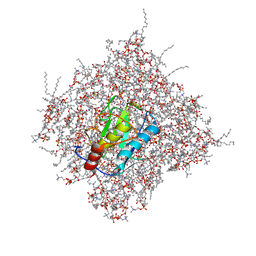 | | Membrane Bilayer complex with Matrix Metalloproteinase-12 at its Beta-face | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, Macrophage metalloelastase, ... | | Authors: | Koppisetti, R.K, Fulcher, Y.G, Prior, S.H, Lenoir, M, Overduin, M, Van Doren, S.R. | | Deposit date: | 2014-03-04 | | Release date: | 2014-12-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Ambidextrous binding of cell and membrane bilayers by soluble matrix metalloproteinase-12.
Nat Commun, 5, 2014
|
|
6CLZ
 
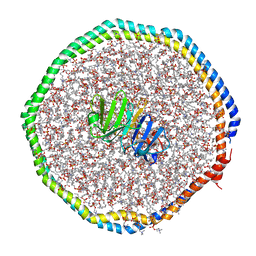 | |
6CM1
 
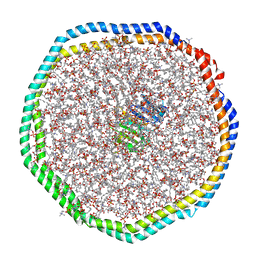 | |
2MZH
 
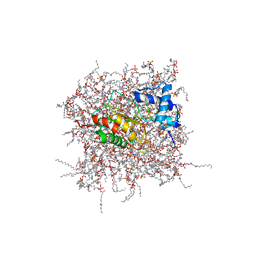 | | NMR Solution Structure of the PRO Form of Human Matrilysin (proMMP-7) in Complex with Zwitterionic Membrane | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, Matrilysin, ... | | Authors: | Prior, S.H, Van Doren, S.R. | | Deposit date: | 2015-02-12 | | Release date: | 2015-11-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Charge-Triggered Membrane Insertion of Matrix Metalloproteinase-7, Supporter of Innate Immunity and Tumors.
Structure, 23, 2015
|
|
2MZI
 
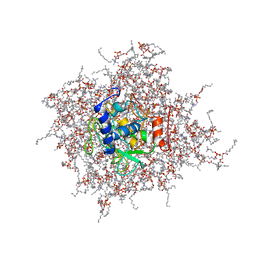 | | NMR Solution Structure of the PRO Form of Human Matrilysin (proMMP-7) in Complex with Anionic Membrane | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, CHOLEST-5-EN-3-YL HYDROGEN SULFATE, ... | | Authors: | Prior, S.H, Van Doren, S.R. | | Deposit date: | 2015-02-12 | | Release date: | 2016-09-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Charge-Triggered Membrane Insertion of Matrix Metalloproteinase-7, Supporter of Innate Immunity and Tumors.
Structure, 23, 2015
|
|
