1QKJ
 
 | | T4 Phage B-Glucosyltransferase, Substrate Binding and Proposed Catalytic Mechanism | | Descriptor: | BETA-GLUCOSYLTRANSFERASE, URIDINE-5'-DIPHOSPHATE | | Authors: | Morera, S, Imberty, I, Aschke-Sonnenborn, U, Ruger, W, Freemont, P.S. | | Deposit date: | 1999-07-22 | | Release date: | 1999-07-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | T4 Phage Beta-Glucosyltransferase: Substrate Binding and Proposed Catalytic Mechanism
J.Mol.Biol., 292, 1999
|
|
2BGU
 
 | | CRYSTAL STRUCTURE OF THE DNA MODIFYING ENZYME BETA-GLUCOSYLTRANSFERASE IN THE PRESENCE AND ABSENCE OF THE SUBSTRATE URIDINE DIPHOSPHOGLUCOSE | | Descriptor: | BETA-GLUCOSYLTRANSFERASE, URIDINE-5'-DIPHOSPHATE | | Authors: | Vrielink, A, Rueger, W, Driessen, H.P.C, Freemont, P.S. | | Deposit date: | 1994-06-09 | | Release date: | 1995-12-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the DNA modifying enzyme beta-glucosyltransferase in the presence and absence of the substrate uridine diphosphoglucose.
EMBO J., 13, 1994
|
|
2BGT
 
 | |
1POS
 
 | | CRYSTAL STRUCTURE OF A NOVEL DISULFIDE-LINKED "TREFOIL" MOTIF FOUND IN A LARGE FAMILY OF PUTATIVE GROWTH FACTORS | | Descriptor: | PORCINE PANCREATIC SPASMOLYTIC POLYPEPTIDE | | Authors: | De, A, Brown, D, Gorman, M, Carr, M, Sanderson, M.R, Freemont, P.S. | | Deposit date: | 1993-10-08 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a disulfide-linked "trefoil" motif found in a large family of putative growth factors.
Proc.Natl.Acad.Sci.USA, 91, 1994
|
|
5N8N
 
 | | Contracted sheath of a Pseudomonas aeruginosa type six secretion system consisting of TssB1 and TssC1 | | Descriptor: | EvpB family type VI secretion protein, Type VI secretion protein, family | | Authors: | Salih, O, He, S, Stach, L, Macdonald, J.T, Planamente, S, Manoli, E, Scheres, S, Filloux, A, Freemont, P.S. | | Deposit date: | 2017-02-23 | | Release date: | 2018-01-10 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Atomic Structure of Type VI Contractile Sheath from Pseudomonas aeruginosa.
Structure, 26, 2018
|
|
4UQZ
 
 | | Coevolution of the ATPase ClpV, the TssB-TssC Sheath and the Accessory HsiE Protein Distinguishes Two Type VI Secretion Classes | | Descriptor: | ACETATE ION, HSIB1, HSIE1 | | Authors: | Forster, A, Planamente, S, Manoli, E, Lossi, N.S, Freemont, P.S, Filloux, A. | | Deposit date: | 2014-06-25 | | Release date: | 2014-10-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Coevolution of the ATPase Clpv, the Sheath Proteins Tssb and Tssc and the Accessory Protein Tagj/Hsie1 Distinguishes Type Vi Secretion Classes.
RNA, 20, 2014
|
|
4UQX
 
 | | Coevolution of the ATPase ClpV, the TssB-TssC Sheath and the Accessory HsiE Protein Distinguishes Two Type VI Secretion Classes | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, HSIE1 | | Authors: | Forster, A, Planamente, S, Manoli, E, Lossi, N.S, Freemont, P.S, Filloux, A. | | Deposit date: | 2014-06-25 | | Release date: | 2014-10-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Coevolution of the ATPase Clpv, the Sheath Proteins Tssb and Tssc and the Accessory Protein Tagj/Hsie1 Distinguishes Type Vi Secretion Classes.
J.Biol.Chem., 289, 2014
|
|
4UQW
 
 | | Coevolution of the ATPase ClpV, the TssB-TssC Sheath and the Accessory HsiE Protein Distinguishes Two Type VI Secretion Classes | | Descriptor: | BENZAMIDINE, PROTEIN CLPV1 | | Authors: | Forster, A, Planamente, S, Manoli, E, Lossi, N.S, Freemont, P.S, Filloux, A. | | Deposit date: | 2014-06-25 | | Release date: | 2014-10-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Coevolution of the ATPase Clpv, the Sheath Proteins Tssb and Tssc and the Accessory Protein Tagj/Hsie1 Distinguishes Type Vi Secretion Classes.
J.Biol.Chem., 289, 2014
|
|
6RRS
 
 | | T=3 MS2 Virus-like particle | | Descriptor: | Capsid protein | | Authors: | de Martin Garrido, N, Ramlaul, K, Simpson, P.A, Crone, M.A, Freemont, P.S, Aylett, C.H.S. | | Deposit date: | 2019-05-20 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Bacteriophage MS2 displays unreported capsid variability assembling T = 4 and mixed capsids.
Mol.Microbiol., 113, 2020
|
|
6RRT
 
 | | T=4 MS2 Virus-like-particle | | Descriptor: | Capsid protein | | Authors: | de Martin Garrido, N, Ramlaul, K, Simpson, P.A, Crone, M.A, Freemont, P.S, Aylett, C.H.S. | | Deposit date: | 2019-05-20 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Bacteriophage MS2 displays unreported capsid variability assembling T = 4 and mixed capsids.
Mol.Microbiol., 113, 2020
|
|
1BGU
 
 | | CRYSTAL STRUCTURE OF THE DNA MODIFYING ENZYME BETA-GLUCOSYLTRANSFERASE IN THE PRESENCE AND ABSENCE OF THE SUBSTRATE URIDINE DIPHOSPHOGLUCOSE | | Descriptor: | BETA-GLUCOSYLTRANSFERASE, URIDINE-5'-DIPHOSPHATE | | Authors: | Vrielink, A, Rueger, W, Driessen, H.P.C, Freemont, P.S. | | Deposit date: | 1994-06-09 | | Release date: | 1994-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the DNA modifying enzyme beta-glucosyltransferase in the presence and absence of the substrate uridine diphosphoglucose.
EMBO J., 13, 1994
|
|
1BGT
 
 | |
1C3J
 
 | | T4 PHAGE BETA-GLUCOSYLTRANSFERASE: SUBSTRATE BINDING AND PROPOSED CATALYTIC MECHANISM | | Descriptor: | BETA-GLUCOSYLTRANSFERASE, URIDINE-5'-DIPHOSPHATE | | Authors: | Morera, S, Imberty, A, Aschke-Sonnenborn, U, Ruger, W, Freemont, P.S. | | Deposit date: | 1999-07-28 | | Release date: | 1999-08-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | T4 phage beta-glucosyltransferase: substrate binding and proposed catalytic mechanism.
J.Mol.Biol., 292, 1999
|
|
1BOR
 
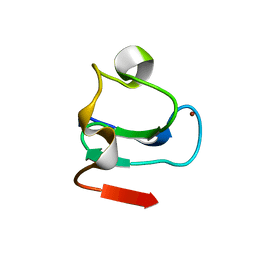 | | TRANSCRIPTION FACTOR PML, A PROTO-ONCOPROTEIN, NMR, 1 REPRESENTATIVE STRUCTURE AT PH 7.5, 30 C, IN THE PRESENCE OF ZINC | | Descriptor: | TRANSCRIPTION FACTOR PML, ZINC ION | | Authors: | Borden, K.L.B, Freemont, P.S. | | Deposit date: | 1995-09-27 | | Release date: | 1997-04-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the RING finger domain from the acute promyelocytic leukaemia proto-oncoprotein PML.
EMBO J., 14, 1995
|
|
1FRE
 
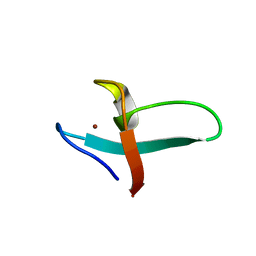 | | XNF7 BBOX, DEVELOPMENTAL PROTEIN, PH 7.5, 30 C, WITH ZINC, NMR, 1 STRUCTURE | | Descriptor: | NUCLEAR FACTOR XNF7, ZINC ION | | Authors: | Borden, K.L.B, Freemont, P.S. | | Deposit date: | 1996-01-31 | | Release date: | 1997-02-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Novel topology of a zinc-binding domain from a protein involved in regulating early Xenopus development.
EMBO J., 14, 1995
|
|
4UQY
 
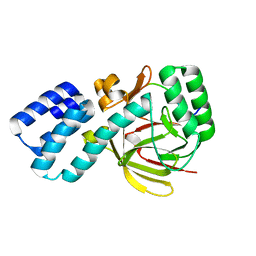 | | Coevolution of the ATPase ClpV, the TssB-TssC Sheath and the Accessory HsiE Protein Distinguishes Two Type VI Secretion Classes | | Descriptor: | HSIB1, HSIE1 | | Authors: | Forster, A, Planamente, S, Manoli, E, Lossi, N.S, Freemont, P.S, Filloux, A. | | Deposit date: | 2014-06-25 | | Release date: | 2014-10-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Coevolution of the ATPase Clpv, the Sheath Proteins Tssb and Tssc and the Accessory Protein Tagj/Hsie1 Distinguishes Type Vi Secretion Classes.
J.Biol.Chem., 289, 2014
|
|
5IIP
 
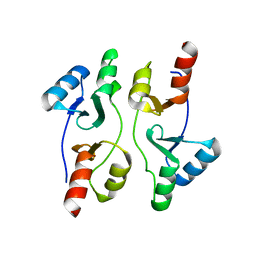 | | Staphylococcus aureus OpuCA | | Descriptor: | Glycine betaine/carnitine/choline ABC transporter%2C ATP-binding protein%2C putative | | Authors: | Tosi, T, Campeotto, I, Freemont, P.S, Grundling, A. | | Deposit date: | 2016-03-01 | | Release date: | 2016-08-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The second messenger c-di-AMP inhibits the osmolyte uptake system OpuC in Staphylococcus aureus.
Sci.Signal., 9, 2016
|
|
6EOW
 
 | |
1SM3
 
 | | CRYSTAL STRUCTURE OF THE TUMOR SPECIFIC ANTIBODY SM3 COMPLEX WITH ITS PEPTIDE EPITOPE | | Descriptor: | CADMIUM ION, CHLORIDE ION, PEPTIDE EPITOPE, ... | | Authors: | Dokurno, P, Bates, P.A, Band, H.A, Stewart, L.M.D, Lally, J.M, Burchell, J.M, Taylor-Papadimitriou, J, Sternberg, M.J.E, Snary, D, Freemont, P.S. | | Deposit date: | 1997-12-23 | | Release date: | 1999-03-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure at 1.95 A resolution of the breast tumour-specific antibody SM3 complexed with its peptide epitope reveals novel hypervariable loop recognition.
J.Mol.Biol., 284, 1998
|
|
3NS9
 
 | | Crystal structure of CDK2 in complex with inhibitor BS-194 | | Descriptor: | (2S,3S)-3-{[7-(benzylamino)-3-(1-methylethyl)pyrazolo[1,5-a]pyrimidin-5-yl]amino}butane-1,2,4-triol, Cell division protein kinase 2 | | Authors: | Hazel, P, Freemont, P.S. | | Deposit date: | 2010-07-01 | | Release date: | 2010-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | A Novel Pyrazolo[1,5-a]pyrimidine Is a Potent Inhibitor of Cyclin-Dependent Protein Kinases 1, 2, and 9, Which Demonstrates Antitumor Effects in Human Tumor Xenografts Following Oral Administration.
J.Med.Chem., 53, 2010
|
|
5JQ5
 
 | | Crystal structure of CDK2 in complex with inhibitor ICEC0942 | | Descriptor: | (3R,4R)-4-[[[7-[(phenylmethyl)amino]-3-propan-2-yl-pyrazolo[1,5-a]pyrimidin-5-yl]amino]methyl]piperidin-3-ol, ACETATE ION, Cyclin-dependent kinase 2 | | Authors: | Hazel, P, Freemont, P.S. | | Deposit date: | 2016-05-04 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Inhibitor Selectivity for Cyclin-Dependent Kinase 7: A Structural, Thermodynamic, and Modelling Study.
ChemMedChem, 12, 2017
|
|
5JQ8
 
 | | Crystal structure of CDK2 in complex with inhibitor ICEC0943 | | Descriptor: | (3S,4S)-4-[[[7-[(phenylmethyl)amino]-3-propan-2-yl-pyrazolo[1,5-a]pyrimidin-5-yl]amino]methyl]piperidin-3-ol, Cyclin-dependent kinase 2 | | Authors: | Hazel, P, Freemont, P.S. | | Deposit date: | 2016-05-04 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Inhibitor Selectivity for Cyclin-Dependent Kinase 7: A Structural, Thermodynamic, and Modelling Study.
ChemMedChem, 12, 2017
|
|
1JRU
 
 | | NMR STRUCTURE OF THE UBX DOMAIN FROM P47 (ENERGY MINIMISED AVERAGE) | | Descriptor: | p47 protein | | Authors: | Yuan, X.M, Shaw, A, Zhang, X.D, Kondo, H, Lally, J, Freemont, P.S, Matthews, S.J. | | Deposit date: | 2001-08-15 | | Release date: | 2001-08-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and interaction surface of the C-terminal domain from p47: a major p97-cofactor involved in SNARE disassembly.
J.Mol.Biol., 311, 2001
|
|
1JIU
 
 | | T4 Phage BGT in Complex with Mg2+ : Form I | | Descriptor: | DNA BETA-GLUCOSYLTRANSFERASE, MAGNESIUM ION, URIDINE-5'-DIPHOSPHATE | | Authors: | Morera, S, Lariviere, L, Kurzeck, J, Aschke-Sonnenborn, U, Freemont, P.S, Janin, J, Ruger, W. | | Deposit date: | 2001-07-03 | | Release date: | 2001-08-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | High resolution crystal structures of T4 phage beta-glucosyltransferase: induced fit and effect of substrate and metal binding.
J.Mol.Biol., 311, 2001
|
|
1JIX
 
 | | T4 Phage BGT in Complex with Ca2+ | | Descriptor: | CALCIUM ION, DNA BETA-GLUCOSYLTRANSFERASE, URIDINE-5'-DIPHOSPHATE | | Authors: | Morera, S, Lariviere, L, Kurzeck, J, Aschke-Sonnenborn, U, Freemont, P.S, Janin, J, Ruger, W. | | Deposit date: | 2001-07-03 | | Release date: | 2001-08-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | High resolution crystal structures of T4 phage beta-glucosyltransferase: induced fit and effect of substrate and metal binding.
J.Mol.Biol., 311, 2001
|
|
