3VF1
 
 | | Structure of a calcium-dependent 11R-lipoxygenase suggests a mechanism for Ca-regulation | | Descriptor: | 11R-lipoxygenase, FE (II) ION, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Eek, P, Jarving, R, Jarving, I, Gilbert, N.C, Newcomer, M.E, Samel, N. | | Deposit date: | 2012-01-09 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.473 Å) | | Cite: | Structure of a Calcium-dependent 11R-Lipoxygenase Suggests a Mechanism for Ca2+ Regulation.
J.Biol.Chem., 287, 2012
|
|
6K35
 
 | | Crystal structure of GH20 exo beta-N-acetylglucosaminidase from Vibrio harveyi in complex with NAG-thiazoline | | Descriptor: | 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, Beta-N-acetylglucosaminidase Nag2 | | Authors: | Meekrathok, P, Stubbs, K.A, Bulmer, D.M, van den Berg, B, Suginta, W. | | Deposit date: | 2019-05-16 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | NAG-thiazoline is a potent inhibitor of the Vibrio campbellii GH20 beta-N-Acetylglucosaminidase.
Febs J., 287, 2020
|
|
7SSA
 
 | | Cryo-EM structure of pioneer factor Cbf1 bound to the nucleosome | | Descriptor: | Centromere-binding protein 1, DNA (137-MER), Histone H2A.1, ... | | Authors: | Eek, P, Tan, S. | | Deposit date: | 2021-11-10 | | Release date: | 2023-04-05 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Basic helix-loop-helix pioneer factors interact with the histone octamer to invade nucleosomes and generate nucleosome-depleted regions.
Mol.Cell, 83, 2023
|
|
6R3R
 
 | | First crystal structure of endo-levanase BT1760 from Bacteroides thetaiotaomicron | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | Eek, P, Ernits, K, Lukk, T, Alamae, T. | | Deposit date: | 2019-03-21 | | Release date: | 2019-06-19 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | First crystal structure of an endo-levanase - the BT1760 from a human gut commensal Bacteroides thetaiotaomicron.
Sci Rep, 9, 2019
|
|
6R3U
 
 | | Endo-levanase BT1760 mutant E221A from Bacteroides thetaiotaomicron complexed with levantetraose | | Descriptor: | GLYCEROL, Glycoside hydrolase family 32, ZINC ION, ... | | Authors: | Eek, P, Ernits, K, Lukk, T, Alamae, T. | | Deposit date: | 2019-03-21 | | Release date: | 2019-06-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | First crystal structure of an endo-levanase - the BT1760 from a human gut commensal Bacteroides thetaiotaomicron.
Sci Rep, 9, 2019
|
|
6EZT
 
 | | Crystal structure of GH20 Exo beta-N-Acetylglucosaminidase D437A inactive mutant from Vibrio harveyi | | Descriptor: | Beta-N-acetylglucosaminidase Nag2, DI(HYDROXYETHYL)ETHER, TETRAETHYLENE GLYCOL | | Authors: | Porfetye, A.T, Meekrathok, P, Burger, M, Vetter, I.R, Suginta, W. | | Deposit date: | 2017-11-16 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of GH20 Exo beta-N-Acetylglucosaminidase from Vibrio harveyi
To Be Published
|
|
6EZR
 
 | |
6EZS
 
 | | Crystal structure of GH20 Exo beta-N-Acetylglucosaminidase from Vibrio harveyi in complex with N-acetylglucosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-N-acetylglucosaminidase Nag2, MALONATE ION | | Authors: | Porfetye, A.T, Meekrathok, P, Burger, M, Vetter, I.R, Suginta, W. | | Deposit date: | 2017-11-16 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of GH20 Exo beta-N-Acetylglucosaminidase from Vibrio harveyi
To Be Published
|
|
6VYP
 
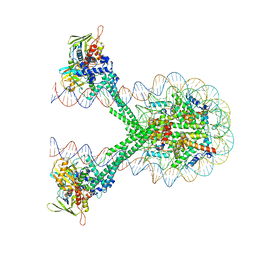 | | Crystal structure of the LSD1/CoREST histone demethylase bound to its nucleosome substrate | | Descriptor: | DNA (191-MER), FLAVIN-ADENINE DINUCLEOTIDE, Histone H2A type 1, ... | | Authors: | Kim, S, Zhu, J, Eek, P, Yennawar, N, Song, T. | | Deposit date: | 2020-02-27 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.99 Å) | | Cite: | Crystal Structure of the LSD1/CoREST Histone Demethylase Bound to Its Nucleosome Substrate.
Mol.Cell, 78, 2020
|
|
1HTE
 
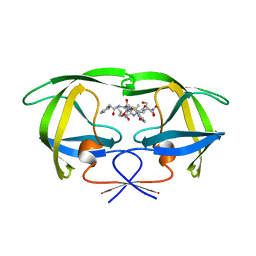 | | X-RAY CRYSTALLOGRAPHIC STUDIES OF A SERIES OF PENICILLIN-DERIVED ASYMMETRIC INHIBITORS OF HIV-1 PROTEASE | | Descriptor: | (2R,4S)-2-[(R)-BENZYLCARBAMOYL-PHENYLACETYL-METHYL]-5,5-DIMETHYL-THIAZOLIDINE-4-CARBOXYLIC ACID, HIV-1 PROTEASE | | Authors: | Jhoti, H, Wonacott, A, Murray-Rust, P. | | Deposit date: | 1994-04-29 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray crystallographic studies of a series of penicillin-derived asymmetric inhibitors of HIV-1 protease.
Biochemistry, 33, 1994
|
|
1HTF
 
 | |
1HTG
 
 | |
