3CHM
 
 | |
4NO4
 
 | | Crystal Structure of Galectin-1 L11A mutant | | Descriptor: | GLYCEROL, Galectin-1, MAGNESIUM ION, ... | | Authors: | Dessau, M. | | Deposit date: | 2013-11-19 | | Release date: | 2014-12-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | Crystal Structure of Galectin-1 L11A mutant
To be Published
|
|
4HJ1
 
 | |
4HJC
 
 | |
6Y9M
 
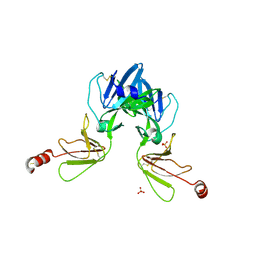 | | Crystal structure of TSWV glycoprotein N ectodomain (sGn) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein, PHOSPHATE ION, ... | | Authors: | Dessau, M, Bahat, Y. | | Deposit date: | 2020-03-10 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structure of tomato spotted wilt virus G N reveals a dimer complex formation and evolutionary link to animal-infecting viruses
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6YA0
 
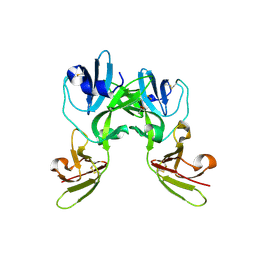 | | Crystal structure of TSWV glycoprotein N ectodomain (Trypsin treated) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, AMMONIUM ION, ... | | Authors: | Dessau, M, Bahat, Y. | | Deposit date: | 2020-03-11 | | Release date: | 2020-09-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Crystal structure of tomato spotted wilt virus G N reveals a dimer complex formation and evolutionary link to animal-infecting viruses
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6YA2
 
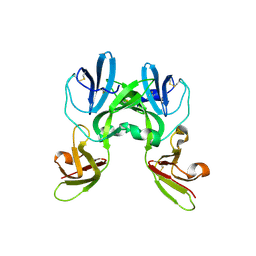 | | Crystal structure of TSWV glycoprotein N ectodomain (Trypsin treated) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein | | Authors: | Dessau, M, Bahat, Y. | | Deposit date: | 2020-03-11 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of tomato spotted wilt virus G N reveals a dimer complex formation and evolutionary link to animal-infecting viruses
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6Y9L
 
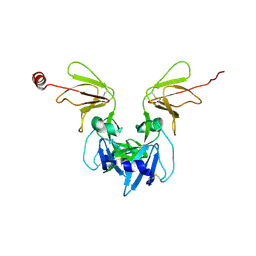 | | Crystal structure of TSWV glycoprotein N ectodomain (sGn) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein, ... | | Authors: | Dessau, M, Bahat, Y. | | Deposit date: | 2020-03-09 | | Release date: | 2020-09-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Crystal structure of tomato spotted wilt virus G N reveals a dimer complex formation and evolutionary link to animal-infecting viruses
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8C6V
 
 | |
6ZG1
 
 | | SARM1 SAM1-2 domains | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sporny, M, Guez-Haddad, J, Khazma, T, Yaron, A, Dessau, M, Mim, C, Isupov, M.N, Zalk, R, Hons, M, Opatowsky, Y. | | Deposit date: | 2020-06-18 | | Release date: | 2020-11-11 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Structural basis for SARM1 inhibition and activation under energetic stress.
Elife, 9, 2020
|
|
6ZFX
 
 | | hSARM1 GraFix-ed | | Descriptor: | (~{E})-4-methylnon-4-enedial, NAD(+) hydrolase SARM1 | | Authors: | Sporny, M, Guez-Haddad, J, Khazma, T, Yaron, A, Dessau, M, Mim, C, Isupov, M.N, Zalk, R, Hons, M, Opatowsky, Y. | | Deposit date: | 2020-06-18 | | Release date: | 2020-11-18 | | Last modified: | 2022-11-09 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Structural basis for SARM1 inhibition and activation under energetic stress.
Elife, 9, 2020
|
|
8QRJ
 
 | | LCC-ICCG PETase mutant H218Y | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, CALCIUM ION, ... | | Authors: | Orr, G, Niv, Y, Barakat, M, Boginya, A, Dessau, M, Afriat-Jurnou, L. | | Deposit date: | 2023-10-09 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Streamlined screening of extracellularly expressed PETase libraries for improved polyethylene terephthalate degradation.
Biotechnol J, 19, 2024
|
|
5J81
 
 | | Crystal structure of Glycoprotein C from Puumala virus in the post-fusion conformation (pH 6.0) | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, ACETATE ION, Envelopment polyprotein, ... | | Authors: | Willensky, S, Dessau, M. | | Deposit date: | 2016-04-07 | | Release date: | 2016-10-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Glycoprotein C from a Hantavirus in the Post-fusion Conformation.
Plos Pathog., 12, 2016
|
|
7ANW
 
 | | hSARM1 NAD+ complex | | Descriptor: | NAD(+) hydrolase SARM1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Sporny, M, Guez-Haddad, J, Khazma, T, Yaron, A, Mim, C, Isupov, M.N, Zalk, R, Dessau, M, Hons, M, Opatowsky, Y. | | Deposit date: | 2020-10-13 | | Release date: | 2020-11-11 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Structural basis for SARM1 inhibition and activation under energetic stress.
Elife, 9, 2020
|
|
8AX4
 
 | |
8AXF
 
 | | Crystal structure of FMV N bound to 42-mer ssRNA | | Descriptor: | GLYCEROL, MAGNESIUM ION, Nucleocapsid protein, ... | | Authors: | Izhaki-Tavor, L, Yechezkel, I, Dessau, M. | | Deposit date: | 2022-08-31 | | Release date: | 2023-04-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | RNA Encapsulation Mode and Evolutionary Insights from the Crystal Structure of Emaravirus Nucleoprotein.
Microbiol Spectr, 11, 2023
|
|
5J9H
 
 | |
6ZG0
 
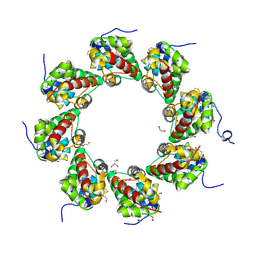 | | SARM1 SAM1-2 domains | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sporny, M, Guez-Haddad, J, Khazma, T, Yaron, A, Dessau, M, Mim, C, Isupov, M.N, Zalk, R, Hons, M, Opatowsky, Y. | | Deposit date: | 2020-06-18 | | Release date: | 2020-11-11 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Structural basis for SARM1 inhibition and activation under energetic stress.
Elife, 9, 2020
|
|
5NVJ
 
 | |
5NXJ
 
 | |
5NV1
 
 | |
4DQ7
 
 | |
4DQJ
 
 | | Structural Investigation of Bacteriophage Phi6 Lysin (in complex with chitotetraose) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, Membrane protein Phi6 P5 | | Authors: | Dessau, M.A, Modis, Y. | | Deposit date: | 2012-02-16 | | Release date: | 2013-01-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Selective pressure causes an RNA virus to trade reproductive fitness for increased structural and thermal stability of a viral enzyme.
PLoS Genet, 8, 2012
|
|
4DQ5
 
 | | Structural Investigation of Bacteriophage Phi6 Lysin (WT) | | Descriptor: | 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, Membrane protein Phi6 P5wt | | Authors: | Dessau, M.A, Modis, Y. | | Deposit date: | 2012-02-15 | | Release date: | 2013-01-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.395 Å) | | Cite: | Selective pressure causes an RNA virus to trade reproductive fitness for increased structural and thermal stability of a viral enzyme.
PLoS Genet, 8, 2012
|
|
3G7T
 
 | |
