6LFQ
 
 | | Crystal structure of Poa1p in apo form | | Descriptor: | ADP-ribose 1''-phosphate phosphatase, GLYCEROL | | Authors: | Chiu, Y.C, Hsu, C.H. | | Deposit date: | 2019-12-03 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.359 Å) | | Cite: | Expanding the Substrate Specificity of Macro Domains toward 3''-Isomer of O-Acetyl-ADP-ribose
Acs Catalysis, 11, 2021
|
|
6LFR
 
 | | Poa1p in complex with ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, ADP-ribose 1''-phosphate phosphatase | | Authors: | Chiu, Y.C, Hsu, C.H. | | Deposit date: | 2019-12-03 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Expanding the Substrate Specificity of Macro Domains toward 3''-Isomer of O-Acetyl-ADP-ribose
Acs Catalysis, 11, 2021
|
|
6LFU
 
 | | Poa1p F152A mutant in complex with ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, ADP-ribose 1''-phosphate phosphatase | | Authors: | Chiu, Y.C, Hsu, C.H. | | Deposit date: | 2019-12-03 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.123 Å) | | Cite: | Expanding the Substrate Specificity of Macro Domains toward 3''-Isomer of O-Acetyl-ADP-ribose
Acs Catalysis, 11, 2021
|
|
6LFT
 
 | | Poa1p S30A mutant in complex with ADP-ribose | | Descriptor: | ACETATE ION, ADENOSINE-5-DIPHOSPHORIBOSE, ADP-ribose 1''-phosphate phosphatase | | Authors: | Chiu, Y.C, Hsu, C.H. | | Deposit date: | 2019-12-03 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Expanding the Substrate Specificity of Macro Domains toward 3''-Isomer of O-Acetyl-ADP-ribose
Acs Catalysis, 11, 2021
|
|
6LFS
 
 | | Poa1p H23A mutant in complex with ADP-ribose | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, ADENOSINE-5-DIPHOSPHORIBOSE, ADP-ribose 1''-phosphate phosphatase | | Authors: | Chiu, Y.C, Hsu, C.H. | | Deposit date: | 2019-12-03 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Expanding the Substrate Specificity of Macro Domains toward 3''-Isomer of O-Acetyl-ADP-ribose
Acs Catalysis, 11, 2021
|
|
2CWV
 
 | | Product schiff-base intermediate of copper amine oxidase from arthrobacter globiformis | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase | | Authors: | Chiu, Y.C, Okajima, T, Murakawa, T, Uchida, M, Taki, M, Hirota, S, Kim, M, Yamaguchi, H, Kawano, Y, Kamiya, N, Kuroda, S, Hayashi, H, Yamamoto, Y, Tanizawa, K. | | Deposit date: | 2005-06-26 | | Release date: | 2006-05-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Kinetic and Structural Studies on the Catalytic Role of the Aspartic Acid Residue Conserved in Copper Amine Oxidase(,)
Biochemistry, 45, 2006
|
|
2CWU
 
 | | Substrate schiff-base intermediate of copper amine oxidase from arthrobacter globiformis | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase | | Authors: | Chiu, Y.C, Okajima, T, Murakawa, T, Uchida, M, Taki, M, Hirota, S, Kim, M, Yamaguchi, H, Kawano, Y, Kamiya, N, Kuroda, S, Hayashi, H, Yamamoto, Y, Tanizawa, K. | | Deposit date: | 2005-06-26 | | Release date: | 2006-05-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Kinetic and Structural Studies on the Catalytic Role of the Aspartic Acid Residue Conserved in Copper Amine Oxidase(,)
Biochemistry, 45, 2006
|
|
2CWT
 
 | | Catalytic base deletion in copper amine oxidase from arthrobacter globiformis | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase | | Authors: | Chiu, Y.C, Okajima, T, Murakawa, T, Uchida, M, Taki, M, Hirota, S, Kim, M, Yamaguchi, H, Kawano, Y, Kamiya, N, Kuroda, S, Hayashi, H, Yamamoto, Y, Tanizawa, K. | | Deposit date: | 2005-06-26 | | Release date: | 2006-05-02 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Kinetic and Structural Studies on the Catalytic Role of the Aspartic Acid Residue Conserved in Copper Amine Oxidase(,)
Biochemistry, 45, 2006
|
|
7F2V
 
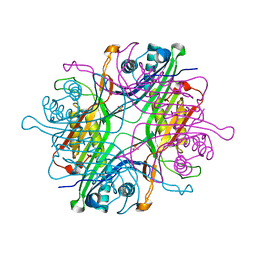 | | Urate oxidase from Thermobispora bispora in apo form | | Descriptor: | DIMETHYL SULFOXIDE, SULFATE ION, Uricase | | Authors: | Chiu, Y.C, Hsu, T.S, Huang, C.Y, Hsu, C.H. | | Deposit date: | 2021-06-14 | | Release date: | 2022-05-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and biochemical insights into a hyperthermostable urate oxidase from Thermobispora bispora for hyperuricemia and gout therapy.
Int.J.Biol.Macromol., 188, 2021
|
|
7F2W
 
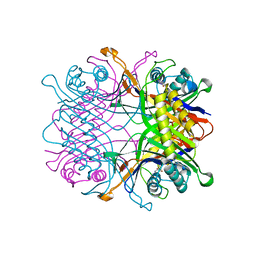 | | TbUox in complex with uric acid | | Descriptor: | URIC ACID, Uricase | | Authors: | Chiu, Y.C, Hsu, T.S, Huang, C.Y, Hsu, C.H. | | Deposit date: | 2021-06-14 | | Release date: | 2022-05-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural and biochemical insights into a hyperthermostable urate oxidase from Thermobispora bispora for hyperuricemia and gout therapy.
Int.J.Biol.Macromol., 188, 2021
|
|
8JBQ
 
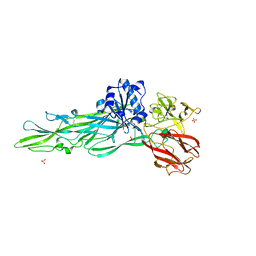 | |
1WMO
 
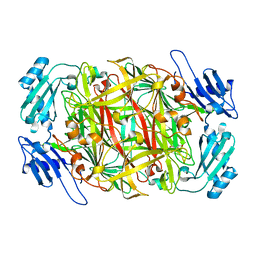 | | Crystal structure of topaquinone-containing amine oxidase activated by nickel ion | | Descriptor: | NICKEL (II) ION, Phenylethylamine oxidase | | Authors: | Okajima, T, Kishishita, S, Chiu, Y.C, Murakawa, T, Kim, M, Yamaguchi, H, Hirota, S, Kuroda, S, Tanizawa, K. | | Deposit date: | 2004-07-13 | | Release date: | 2005-08-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Reinvestigation of metal ion specificity for quinone cofactor biogenesis in bacterial copper amine oxidase
Biochemistry, 44, 2005
|
|
1WMP
 
 | | Crystal structure of amine oxidase complexed with cobalt ion | | Descriptor: | COBALT (II) ION, Phenylethylamine oxidase | | Authors: | Okajima, T, Kishishita, S, Chiu, Y.C, Murakawa, T, Kim, M, Yamaguchi, H, Hirota, S, Kuroda, S, Tanizawa, K. | | Deposit date: | 2004-07-13 | | Release date: | 2005-08-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Reinvestigation of metal ion specificity for quinone cofactor biogenesis in bacterial copper amine oxidase
Biochemistry, 44, 2005
|
|
1WMN
 
 | | Crystal structure of topaquinone-containing amine oxidase activated by cobalt ion | | Descriptor: | COBALT (II) ION, Phenylethylamine oxidase | | Authors: | Okajima, T, Kishishita, S, Chiu, Y.C, Murakawa, T, Kim, M, Yamaguchi, H, Hirota, S, Kuroda, S, Tanizawa, K. | | Deposit date: | 2004-07-13 | | Release date: | 2005-08-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Reinvestigation of metal ion specificity for quinone cofactor biogenesis in bacterial copper amine oxidase
Biochemistry, 44, 2005
|
|
8JC7
 
 | | Cryo-EM structure of Vibrio campbellii alpha-hemolysin | | Descriptor: | CALCIUM ION, Hemolysin, POTASSIUM ION | | Authors: | Wang, C.H, Yeh, M.K, Ho, M.C, Lin, S.M. | | Deposit date: | 2023-05-10 | | Release date: | 2023-09-27 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (2.06 Å) | | Cite: | Structural basis for calcium-stimulating pore formation of Vibrio alpha-hemolysin.
Nat Commun, 14, 2023
|
|
6LXN
 
 | |
6LXM
 
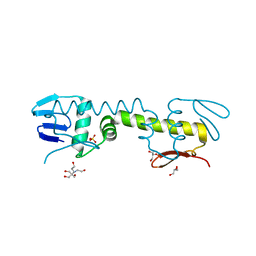 | | Crystal structure of C-terminal DNA-binding domain of Escherichia coli OmpR as a domain-swapped dimer | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, SULFATE ION, ... | | Authors: | Sadotra, S, Chen, C, Hsu, C.H. | | Deposit date: | 2020-02-11 | | Release date: | 2020-12-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.412 Å) | | Cite: | Structural basis for promoter DNA recognition by the response regulator OmpR.
J.Struct.Biol., 213, 2020
|
|
6LXL
 
 | |
7VE6
 
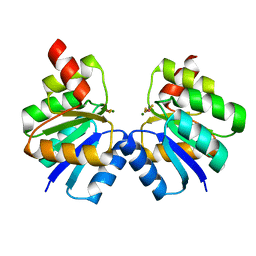 | | N-terminal domain of VraR | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, Response regulator protein VraR | | Authors: | Kumar, J.V, Chen, C, Hsu, C.H. | | Deposit date: | 2021-09-08 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structural insights into DNA binding domain of vancomycin-resistance-associated response regulator in complex with its promoter DNA from Staphylococcus aureus.
Protein Sci., 31, 2022
|
|
7VE4
 
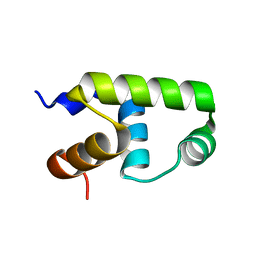 | | C-terminal domain of VraR | | Descriptor: | DNA-binding response regulator | | Authors: | Kumar, J.V, Chen, C, Hsu, C.H. | | Deposit date: | 2021-09-08 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural insights into DNA binding domain of vancomycin-resistance-associated response regulator in complex with its promoter DNA from Staphylococcus aureus.
Protein Sci., 31, 2022
|
|
7VE5
 
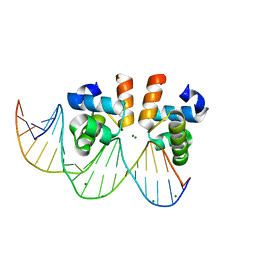 | | C-terminal domain of VraR | | Descriptor: | DNA-binding response regulator, MAGNESIUM ION, R1-DNA | | Authors: | Kumar, J.V, Chen, C, Hsu, C.H. | | Deposit date: | 2021-09-08 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into DNA binding domain of vancomycin-resistance-associated response regulator in complex with its promoter DNA from Staphylococcus aureus.
Protein Sci., 31, 2022
|
|
7C4H
 
 | |
7C33
 
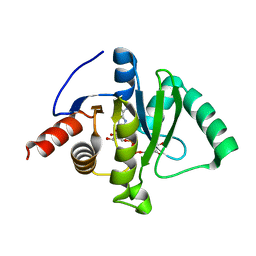 | | Macro domain of SARS-CoV-2 in complex with ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Non-structural protein 3 | | Authors: | Lin, M.H, Hsu, C.H. | | Deposit date: | 2020-05-11 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.83 Å) | | Cite: | Structural, Biophysical, and Biochemical Elucidation of the SARS-CoV-2 Nonstructural Protein 3 Macro Domain.
Acs Infect Dis., 6, 2020
|
|
7CZ4
 
 | |
5ZDA
 
 | |
