4UA1
 
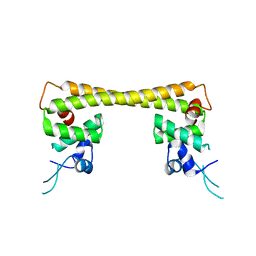 | | Crystal structure of dual function transcriptional regulator MerR form Bacillus megaterium MB1 in complex with mercury (II) ion | | Descriptor: | MERCURY (II) ION, Regulatory protein | | Authors: | Chang, C.C, Lin, L.Y, Zou, X.W, Huang, C.C, Chan, N.L. | | Deposit date: | 2014-08-07 | | Release date: | 2015-07-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structural basis of the mercury(II)-mediated conformational switching of the dual-function transcriptional regulator MerR
Nucleic Acids Res., 43, 2015
|
|
4UA2
 
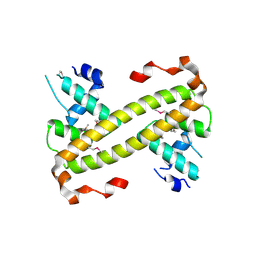 | | Crystal structure of dual function transcriptional regulator MerR from Bacillus megaterium MB1 | | Descriptor: | Regulatory protein | | Authors: | Lin, L.Y, Chang, C.C, Zou, X.W, Huang, C.C, Chan, N.L. | | Deposit date: | 2014-08-07 | | Release date: | 2015-07-22 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural basis of the mercury(II)-mediated conformational switching of the dual-function transcriptional regulator MerR
Nucleic Acids Res., 43, 2015
|
|
7XMV
 
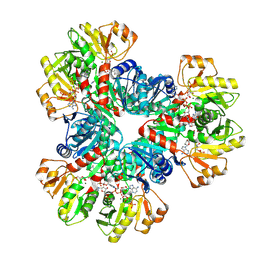 | | E.coli phosphoribosylpyrophosphate (PRPP) synthetase type A(AMP/ADP) filament bound with ADP, AMP and R5P | | Descriptor: | 5-O-phosphono-alpha-D-ribofuranose, ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Hu, H.H, Lu, G.M, Chang, C.C, Liu, J.L. | | Deposit date: | 2022-04-27 | | Release date: | 2022-06-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Filamentation modulates allosteric regulation of PRPS.
Elife, 11, 2022
|
|
7XN3
 
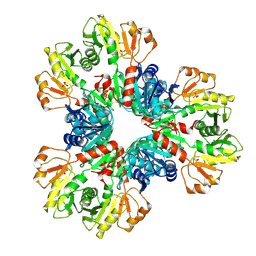 | | E.coli phosphoribosylpyrophosphate (PRPP) synthetase type B filament bound with Pi | | Descriptor: | PHOSPHATE ION, Ribose-phosphate pyrophosphokinase | | Authors: | Hu, H.H, Lu, G.M, Chang, C.C, Liu, J.L. | | Deposit date: | 2022-04-27 | | Release date: | 2022-06-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Filamentation modulates allosteric regulation of PRPS.
Elife, 11, 2022
|
|
7XMU
 
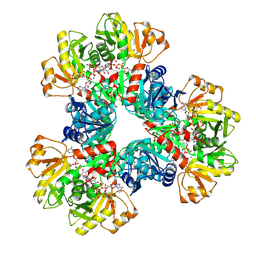 | | E.coli phosphoribosylpyrophosphate (PRPP) synthetase type A filament bound with ADP, Pi and R5P | | Descriptor: | 5-O-phosphono-alpha-D-ribofuranose, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Hu, H.H, Lu, G.M, Chang, C.C, Liu, J.L. | | Deposit date: | 2022-04-26 | | Release date: | 2022-06-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Filamentation modulates allosteric regulation of PRPS.
Elife, 11, 2022
|
|
6L1S
 
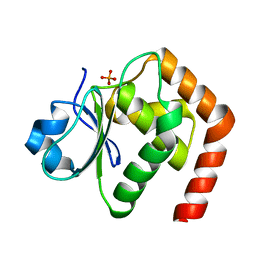 | | Crystal structure of DUSP22 mutant_C88S | | Descriptor: | Dual specificity protein phosphatase 22, PHOSPHATE ION | | Authors: | Lai, C.H, Chang, C.C, Lyu, P.C. | | Deposit date: | 2019-09-30 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.3611 Å) | | Cite: | Structural Insights into the Active Site Formation of DUSP22 in N-loop-containing Protein Tyrosine Phosphatases.
Int J Mol Sci, 21, 2020
|
|
7DPT
 
 | | Structural basis for ligand binding modes of CTP synthase | | Descriptor: | 6-DIAZENYL-5-OXO-L-NORLEUCINE, ADENOSINE-5'-DIPHOSPHATE, CTP synthase, ... | | Authors: | Liu, J.L, Zhou, X, Guo, C.J, Chang, C.C. | | Deposit date: | 2020-12-21 | | Release date: | 2021-09-15 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | Structural basis for ligand binding modes of CTP synthase.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7DPW
 
 | | Structural basis for ligand binding modes of CTP synthase | | Descriptor: | CTP synthase, CYTIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Liu, J.L, Zhou, X, Guo, C.J, Chang, C.C. | | Deposit date: | 2020-12-21 | | Release date: | 2021-09-15 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Structural basis for ligand binding modes of CTP synthase.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4M9O
 
 | |
4M8V
 
 | |
2KQS
 
 | |
6LMY
 
 | | Crystal structure of DUSP22 mutant_C88S/S93A | | Descriptor: | Dual specificity protein phosphatase 22, PHOSPHATE ION | | Authors: | Lai, C.H, Lyu, P.C. | | Deposit date: | 2019-12-27 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Insights into the Active Site Formation of DUSP22 in N-loop-containing Protein Tyrosine Phosphatases.
Int J Mol Sci, 21, 2020
|
|
6LOT
 
 | | Crystal structure of DUSP22 mutant_N128D | | Descriptor: | Dual specificity protein phosphatase 22, SULFATE ION | | Authors: | Lai, C.H, Lyu, P.C. | | Deposit date: | 2020-01-07 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural Insights into the Active Site Formation of DUSP22 in N-loop-containing Protein Tyrosine Phosphatases.
Int J Mol Sci, 21, 2020
|
|
6LFG
 
 | |
6M4T
 
 | | U shaped head to head four-way junction in d(TTCTGCTGCTGAA) sequence | | Descriptor: | COBALT (II) ION, DNA (5'-D(P*(UD)P*TP*CP*TP*GP*CP*TP*GP*CP*TP*GP*AP*A)-3'), N4-[4-[(6-chloranyl-2-methoxy-acridin-9-yl)amino]butyl]-1,3,5-triazine-2,4,6-triamine | | Authors: | Hou, M.H, Chien, C.M, Satange, R.B, Wu, P.C. | | Deposit date: | 2020-03-09 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Basis for Targeting T:T Mismatch with Triaminotriazine-Acridine Conjugate Induces a U-Shaped Head-to-Head Four-Way Junction in CTG Repeat DNA.
J.Am.Chem.Soc., 142, 2020
|
|
6M5J
 
 | | U shaped head to head four-way junction in d(TTCTGCTGCTGAA/TTCTGCAGCTGAA) sequence | | Descriptor: | COBALT (II) ION, DNA (5'-D(P*TP*TP*CP*TP*GP*CP*AP*GP*CP*TP*GP*AP*A)-3'), DNA (5'-D(P*TP*TP*CP*TP*GP*CP*TP*GP*CP*TP*GP*AP*A)-3'), ... | | Authors: | Hou, M.H, Chien, C.M, Satange, R.B. | | Deposit date: | 2020-03-11 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Basis for Targeting T:T Mismatch with Triaminotriazine-Acridine Conjugate Induces a U-Shaped Head-to-Head Four-Way Junction in CTG Repeat DNA.
J.Am.Chem.Soc., 142, 2020
|
|
6LVQ
 
 | | Crystal structure of DUSP22_VO4 | | Descriptor: | Dual specificity protein phosphatase 22, VANADATE ION | | Authors: | Lai, C.H, Lyu, P.C. | | Deposit date: | 2020-02-04 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structural Insights into the Active Site Formation of DUSP22 in N-loop-containing Protein Tyrosine Phosphatases.
Int J Mol Sci, 21, 2020
|
|
6LOU
 
 | | Crystal structure of DUSP22 mutant_C88S/S93N | | Descriptor: | Dual specificity protein phosphatase 22, PHOSPHATE ION | | Authors: | Lai, C.H, Lyu, P.C. | | Deposit date: | 2020-01-07 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5301 Å) | | Cite: | Structural Insights into the Active Site Formation of DUSP22 in N-loop-containing Protein Tyrosine Phosphatases.
Int J Mol Sci, 21, 2020
|
|
8W76
 
 | | Crystal structure of d(CGTATACG)2 duplex | | Descriptor: | DNA (5'-D(P*CP*GP*TP*AP*TP*AP*CP*G)-3'), MANGANESE (II) ION | | Authors: | Satange, R.B, Peng, C.L, Hou, M.H. | | Deposit date: | 2023-08-30 | | Release date: | 2024-08-07 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Targeting DNA junction sites by bis-intercalators induces topological changes with potent antitumor effects.
Nucleic Acids Res., 52, 2024
|
|
8W7W
 
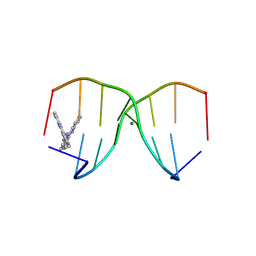 | | Crystal structure of d(CGTATACG)2 with acridine complex | | Descriptor: | DNA (5'-D(P*CP*GP*TP*AP*TP*AP*CP*G)-3'), MAGNESIUM ION, ~{N},~{N}'-di(acridin-9-yl)pentane-1,5-diamine | | Authors: | Huang, S.C, Satange, R.B, Hou, M.H. | | Deposit date: | 2023-08-31 | | Release date: | 2024-08-07 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Targeting DNA junction sites by bis-intercalators induces topological changes with potent antitumor effects.
Nucleic Acids Res., 52, 2024
|
|
8WQ7
 
 | | Crystal structure of d(CGTATACG)2 with a four-carbon linker containing diacridine compound | | Descriptor: | DNA (5'-D(P*CP*GP*TP*AP*TP*AP*CP*G)-3'), MANGANESE (II) ION, N,N'-di(acridin-9-yl)butane-1,4-diamine | | Authors: | Huang, S.C, Satange, R.B, Hou, M.H. | | Deposit date: | 2023-10-11 | | Release date: | 2024-08-07 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Targeting DNA junction sites by bis-intercalators induces topological changes with potent antitumor effects.
Nucleic Acids Res., 52, 2024
|
|
6L47
 
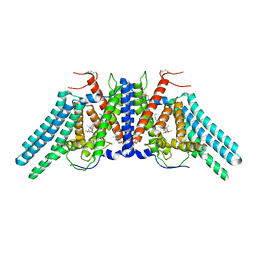 | | Structure of the human sterol O-acyltransferase 1 in complex with CI-976 | | Descriptor: | 2,2-dimethyl-N-(2,4,6-trimethoxyphenyl)dodecanamide, CHOLESTEROL, Sterol O-acyltransferase 1 | | Authors: | Chen, L, Guan, C, Niu, Y. | | Deposit date: | 2019-10-16 | | Release date: | 2020-04-29 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into the inhibition mechanism of human sterol O-acyltransferase 1 by a competitive inhibitor.
Nat Commun, 11, 2020
|
|
7YK1
 
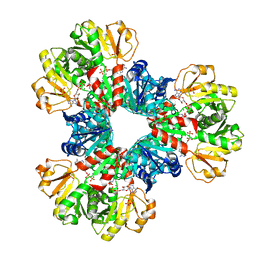 | | Structural basis of human PRPS2 filaments | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Lu, G.M, Hu, H.H, Liu, J.L. | | Deposit date: | 2022-07-21 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structural basis of human PRPS2 filaments.
Cell Biosci, 13, 2023
|
|
4NPN
 
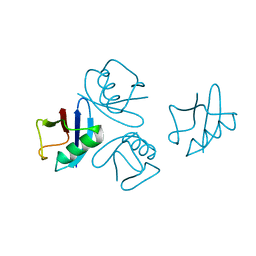 | | Crystal structure of human tetra-SUMO-2 | | Descriptor: | Small ubiquitin-related modifier 2 | | Authors: | Kung, C.C.-H, Naik, M.T, Chen, C.L, Ma, C, Huang, T.H. | | Deposit date: | 2013-11-22 | | Release date: | 2014-10-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.633 Å) | | Cite: | Structural analysis of poly-SUMO chain recognition by the RNF4-SIMs domain.
Biochem.J., 462, 2014
|
|
6AHO
 
 | | Crystal structure of Kap114p | | Descriptor: | Importin subunit beta-5 | | Authors: | Liao, C.C, Shankar, S, Ahmed, G.R, Hsia, K.C. | | Deposit date: | 2018-08-20 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Karyopherin Kap114p-mediated trans-repression controls ribosomal gene expression under saline stress.
Embo Rep., 21, 2020
|
|
